1ZM0
 
 | | Crystal Structure of the Carboxyl Terminal PH Domain of Pleckstrin To 2.1 Angstroms | | Descriptor: | Pleckstrin | | Authors: | Jackson, S.G, Zhang, Y, Zhang, K, Summerfield, R, Haslam, R.J, Junop, M.S. | | Deposit date: | 2005-05-09 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the carboxy-terminal PH domain of pleckstrin at 2.1 Angstroms.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3O95
 
 | | Crystal Structure of Human DPP4 Bound to TAK-100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a 3-Pyridylacetic Acid Derivative (TAK-100) as a Potent, Selective and Orally Active Dipeptidyl Peptidase IV (DPP-4) Inhibitor.
J.Med.Chem., 53, 2011
|
|
3O8I
 
 | | Structure of 14-3-3 isoform sigma in complex with a C-Raf1 peptide and a stabilizing small molecule fragment | | Descriptor: | 14-3-3 binding site peptide of RAF proto-oncogene serine/threonine-protein kinase, 14-3-3 protein sigma, 6,6-dihydroxy-1-methoxyhexan-2-one | | Authors: | Ottmann, C, Rose, R, Kaiser, M, Kuhenne, P. | | Deposit date: | 2010-08-03 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Impaired Binding of 14-3-3 to C-RAF in Noonan Syndrome Suggests New Approaches in Diseases with Increased Ras Signaling
Mol.Cell.Biol., 30, 2010
|
|
3OAK
 
 | | Crystal structure of a Spn1 (Iws1)-Spt6 complex | | Descriptor: | Transcription elongation factor SPT6, Transcription factor IWS1 | | Authors: | McDonald, S.M, Close, D, Hill, C.P. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and biological importance of the spn1-spt6 interaction, and its regulatory role in nucleosome binding.
Mol.Cell, 40, 2010
|
|
2PNK
 
 | |
3OC9
 
 | |
3OA6
 
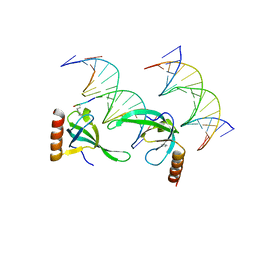 | | Human MSL3 Chromodomain bound to DNA and H4K20me1 peptide | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), H4 peptide monomethylated at lysine 20, ... | | Authors: | Kim, D, Huang, P, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2PRR
 
 | |
3OB1
 
 | |
3OBE
 
 | |
3NOM
 
 | | Crystal Structure of Zymomonas mobilis Glutaminyl Cyclase (monoclinic form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
2PG4
 
 | |
3NTN
 
 | | Crystal Structure of UspA1 head and neck domain from Moraxella catarrhalis | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Conners, R, Zaccai, N, Agnew, C, Burton, N, Brady, R.L. | | Deposit date: | 2010-07-05 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correlation of in situ mechanosensitive responses of the Moraxella catarrhalis adhesin UspA1 with fibronectin and receptor CEACAM1 binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1XJ5
 
 | | X-RAY STRUCTURE OF SPERMIDINE SYNTHASE FROM ARABIDOPSIS THALIANA GENE AT1G23820 | | Descriptor: | Spermidine synthase 1 | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-22 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-RAY STRUCTURE OF SPERMIDINE SYNTHASE FROM ARABIDOPSIS THALIANA GENE AT1G23820
To be published
|
|
3OKN
 
 | | Crystal structure of S25-39 in complex with Kdo(2.4)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
3OM4
 
 | | Crystal structure of B. megaterium levansucrase mutant K373A | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Levansucrase, ... | | Authors: | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of B. megaterium levansucrase mutant K373A
To be Published
|
|
3OMQ
 
 | | Fragment-Based Design of novel Estrogen Receptor Ligands | | Descriptor: | 2-[(trifluoromethyl)sulfonyl]-1,2,3,4-tetrahydroisoquinolin-6-ol, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, van Otterlo, W.A, Rose, R, Fuchs, S, Dominguez Seoane, M, Waldmann, H, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design and Evaluation of Fragment-Like Estrogen Receptor Tetrahydroisoquinoline Ligands from a Scaffold-Detection Approach.
J.Med.Chem., 54, 2011
|
|
1XX7
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-403030-001 | | Descriptor: | NICKEL (II) ION, UNKNOWN ATOM OR ION, oxetanocin-like protein | | Authors: | Chen, L, Tempel, W, Habel, J, Zhou, W, Nguyen, D, Chang, S.-H, Lee, D, Kelley, L.-L.C, Dillard, B.D, Liu, Z.-J, Bridger, S, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-11-04 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Conserved hypothetical protein from Pyrococcus furiosus Pfu-403030-001
To be published
|
|
3OOQ
 
 | | CRYSTAL STRUCTURE OF amidohydrolase from Thermotoga maritima MSB8 | | Descriptor: | GLYCEROL, amidohydrolase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-15 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | CRYSTAL STRUCTURE OF amidohydrolase from Thermotoga maritima MSB8
To be Published
|
|
3OQQ
 
 | |
2OKF
 
 | |
2I8D
 
 | |
3OTE
 
 | |
3OWD
 
 | | Crystal Structure of HSP90 with N-Aryl-benzimidazolone II | | Descriptor: | Heat shock protein HSP 90-alpha, N-{[1-(5-chloro-2,4-dihydroxyphenyl)-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]methyl}naphthalene-1-sulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2O3L
 
 | |
