3C7A
 
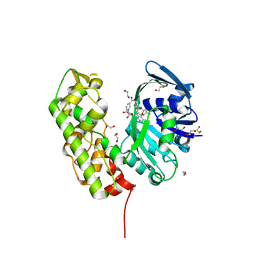 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH) | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | Deposit date: | 2008-02-07 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
1L84
 
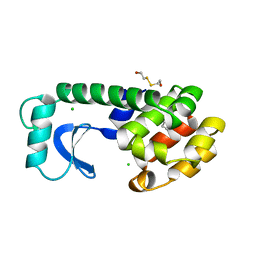 | |
4F04
 
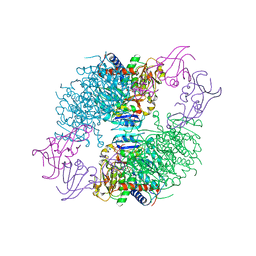 | | A Second Allosteric site in E. coli Aspartate Transcarbamoylase: R-state ATCase with UTP bound | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Peterson, A.W, Cockrell, G.M, Kantrowitz, E.R. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A second allosteric site in Escherichia coli aspartate transcarbamoylase.
Biochemistry, 51, 2012
|
|
4DZL
 
 | | A de novo designed Coiled Coil CC-Tri | | Descriptor: | COILED-COIL PEPTIDE CC-TRI | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
1CSH
 
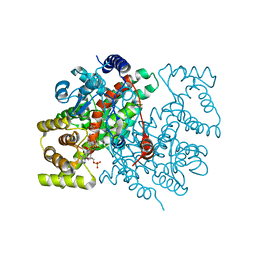 | |
4DZK
 
 | | A de novo designed Coiled Coil CC-Tri-N13 | | Descriptor: | CHLORIDE ION, COILED-COIL PEPTIDE CC-TRI-N13 | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
1L83
 
 | |
3PJP
 
 | | A Tandem SH2 Domain in Transcription Elongation Factor Spt6 Binds the Phosphorylated RNA Polymerase II C-terminal Repeat Domain(CTD) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Transcription elongation factor SPT6 | | Authors: | Sun, M, Lariviere, L, Dengl, S, Mayer, A, Cramer, P. | | Deposit date: | 2010-11-10 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD).
J.Biol.Chem., 285, 2010
|
|
3S0M
 
 | | A Structural Element that Modulates Proton-Coupled Electron Transfer in Oxalate Decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saylor, B.T, Reinhardt, L.A, Lu, Z, Shukla, M.S, Cleland, W.W, Allen, K.N, Richards, N.G.J. | | Deposit date: | 2011-05-13 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A structural element that facilitates proton-coupled electron transfer in oxalate decarboxylase.
Biochemistry, 51, 2012
|
|
1CPF
 
 | | A CATION BINDING MOTIF STABILIZES THE COMPOUND I RADICAL OF CYTOCHROME C PEROXIDASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miller, M.A, Han, G.W, Kraut, J. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cation binding motif stabilizes the compound I radical of cytochrome c peroxidase.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
3M3J
 
 | | A new crystal form of Lys48-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Ubiquitin | | Authors: | Trempe, J.F, Brown, N.R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new crystal form of Lys48-linked diubiquitin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1CPD
 
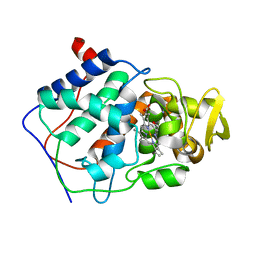 | |
198D
 
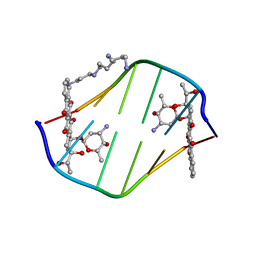 | | A TRIGONAL FORM OF THE IDARUBICIN-D(CGATCG) COMPLEX: CRYSTAL AND MOLECULAR STRUCTURE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), IDARUBICIN, SPERMINE | | Authors: | Dautant, A, Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1994-11-28 | | Release date: | 1995-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A trigonal form of the idarubicin:d(CGATCG) complex; crystal and molecular structure at 2.0 A resolution.
Nucleic Acids Res., 23, 1995
|
|
3Q5U
 
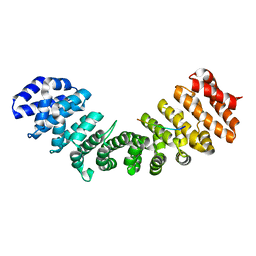 | |
1TC1
 
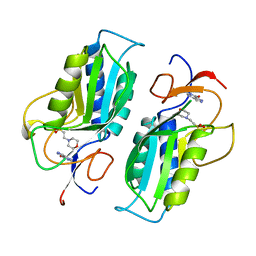 | | A 1.4 ANGSTROM CRYSTAL STRUCTURE FOR THE HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE OF TRYPANOSOMA CRUZI | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMYCIN B, PROTEIN (HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE) | | Authors: | Focia, P.J, Craig III, S.P, Nieves-Alicea, R, Fletterick, R.J, Eakin, A.E. | | Deposit date: | 1998-09-30 | | Release date: | 1999-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A 1.4 A crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi.
Biochemistry, 37, 1998
|
|
1CSI
 
 | |
1CPE
 
 | |
1U8C
 
 | | A novel adaptation of the integrin PSI domain revealed from its crystal structure | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.P, Stehle, T, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2004-08-05 | | Release date: | 2004-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A novel adaptation of the integrin PSI domain revealed from its crystal structure.
J.Biol.Chem., 279, 2004
|
|
1A8W
 
 | |
1B5Q
 
 | | A 30 ANGSTROM U-SHAPED CATALYTIC TUNNEL IN THE CRYSTAL STRUCTURE OF POLYAMINE OXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, N,N'-BIS(2,3-BUTADIENYL)-1,4-BUTANE-DIAMINE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase.
Structure Fold.Des., 7, 1999
|
|
1T31
 
 | | A Dual Inhibitor of the Leukocyte Proteases Cathepsin G and Chymase with Therapeutic Efficacy in Animals Models of Inflammation | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[3-({METHYL[1-(2-NAPHTHOYL)PIPERIDIN-4-YL]AMINO}CARBONYL)-2-NAPHTHYL]-1-(1-NAPHTHYL)-2-OXOETHYLPHOSPHONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | de Garavilla, L, Greco, M.N, Giardino, E.C, Wells, G.I, Haertlein, B.J, Kauffman, J.A, Corcoran, T.W, Derian, C.K, Eckardt, A.J, Abraham, W.M, Sukumar, N, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E, Andrade-Gordon, P, Damiano, B.P, Maryanoff, B.E, Pereira, P.J.B, Wang, Z.M, Rubin, H, Huber, R, Bode, W, Schechter, N.M, Strobl, S. | | Deposit date: | 2004-04-23 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel, potent dual inhibitor of the leukocyte proteases cathepsin G and chymase: molecular mechanisms and anti-inflammatory activity in vivo.
J.Biol.Chem., 280, 2005
|
|
1TGL
 
 | | A SERINE PROTEASE TRIAD FORMS THE CATALYTIC CENTRE OF A TRIACYLGLYCEROL LIPASE | | Descriptor: | TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | Deposit date: | 1990-02-05 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A serine protease triad forms the catalytic centre of a triacylglycerol lipase.
Nature, 343, 1990
|
|
1ADD
 
 | |
2P7D
 
 | | A Minimal, 'Hinged' Hairpin Ribozyme Construct Solved with Mimics of the Product Strands at 2.25 Angstroms Resolution | | Descriptor: | 3' substrate strand, octameric fragment, 5' substrate strand, ... | | Authors: | Torelli, A.T, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization
Rna, 13, 2007
|
|
1B37
 
 | | A 30 ANGSTROM U-SHAPED CATALYTIC TUNNEL IN THE CRYSTAL STRUCTURE OF POLYAMINE OXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (POLYAMINE OXIDASE), ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase.
Structure Fold.Des., 7, 1999
|
|
