3QGT
 
 | | Crystal structure of Wild-type PfDHFR-TS COMPLEXED WITH NADPH, dUMP AND PYRIMETHAMINE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Yuthavong, Y. | | Deposit date: | 2011-01-24 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trypanosomal dihydrofolate reductase reveals natural antifolate resistance
Acs Chem.Biol., 6, 2011
|
|
3QB8
 
 | | Paramecium Chlorella Bursaria Virus1 Putative ORF A654L is a Polyamine Acetyltransferase | | Descriptor: | A654L protein, COENZYME A, IMIDAZOLE | | Authors: | Charlop-Powers, Z, Zhou, M.-M, Jakoncic, J, Gurnon, J, Van Etten, J. | | Deposit date: | 2011-01-12 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Paramecium bursaria chlorella virus 1 encodes a polyamine acetyltransferase.
J. Biol. Chem., 287, 2012
|
|
3QG2
 
 | | Plasmodium falciparum DHFR-TS qradruple mutant (N51I+C59R+S108N+I164L, V1/S) pyrimethamine complex | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Yuvaniyama, J, Taweechai, S, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2011-01-24 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trypanosomal dihydrofolate reductase reveals natural antifolate resistance
Acs Chem.Biol., 6, 2011
|
|
3RI6
 
 | | A Novel Mechanism of Sulfur Transfer Catalyzed by O-Acetylhomoserine Sulfhydrylase in Methionine Biosynthetic Pathway of Wolinella succinogenes | | Descriptor: | O-ACETYLHOMOSERINE SULFHYDRYLASE | | Authors: | Tran, T.H, Krishnamoorthy, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-13 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel mechanism of sulfur transfer catalyzed by O-acetylhomoserine sulfhydrylase in the methionine-biosynthetic pathway of Wolinella succinogenes.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3U0E
 
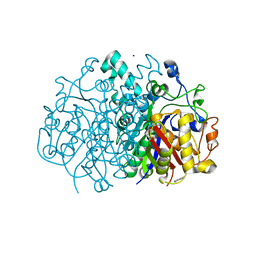 | |
3RR2
 
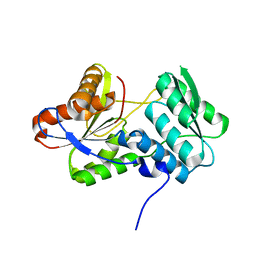 | |
3RBM
 
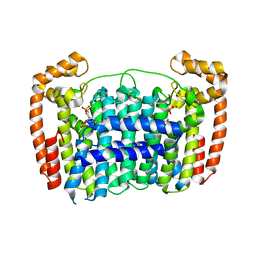 | | Crystal structure of Plasmodium vivax geranylgeranylpyrophosphate synthase complexed with BPH -703 | | Descriptor: | 3-(2,2-diphosphonoethyl)-1-dodecyl-1H-imidazol-3-ium, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Liu, Y.L, No, J.H, Oldfield, E. | | Deposit date: | 2011-03-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Lipophilic analogs of zoledronate and risedronate inhibit Plasmodium geranylgeranyl diphosphate synthase (GGPPS) and exhibit potent antimalarial activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3R96
 
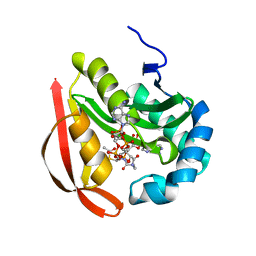 | |
3SGX
 
 | |
3S7Y
 
 | | Crystal structure of mmNAGS in Space Group P3121 at 4.3 A resolution | | Descriptor: | N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.3077 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3U0F
 
 | |
3UBO
 
 | | The crystal structure of adenosine kinase from Sinorhizobium meliloti | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, adenosine kinase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The crystal structure of adenosine kinase from Sinorhizobium meliloti
To be Published
|
|
3TR1
 
 | | Structure of a 3-phosphoshikimate 1-carboxyvinyltransferase (aroA) from Coxiella burnetii | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, PHOSPHATE ION, POTASSIUM ION | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TTQ
 
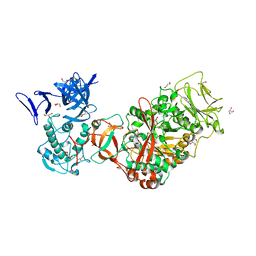 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in orthorhombic apo-form at 1.9 angstrom resolution | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Remaud-Simeon, M, Dijkstra, B.W, Tranier, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
3TMS
 
 | |
3UM5
 
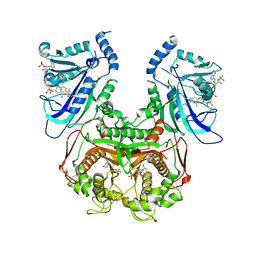 | | Double mutant (A16V+S108T) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS-T9/94) complexed with pyrimethamine, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Kamchonwongpaisan, S, Chitnumsub, P, Yuthavong, Y. | | Deposit date: | 2011-11-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combined Spatial Limitation around Residues 16 and 108 of Plasmodium falciparum Dihydrofolate Reductase Explains Resistance to Cycloguanil.
Antimicrob.Agents Chemother., 56, 2012
|
|
3UM6
 
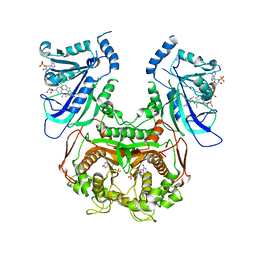 | | Double mutant (A16V+S108T) Plasmodium falciparum DHFR-TS (T9/94) complexed with cycloguanil, NADPH and dUMP | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2011-11-12 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Combined Spatial Limitation around Residues 16 and 108 of Plasmodium falciparum Dihydrofolate Reductase Explains Resistance to Cycloguanil.
Antimicrob.Agents Chemother., 56, 2012
|
|
3UY5
 
 | | crystal structure of Eis from Mycobacterium tuberculosis | | Descriptor: | Enhanced intracellular survival protein | | Authors: | Kim, K.H, Suh, S.W. | | Deposit date: | 2011-12-05 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
3UM8
 
 | | Wild-type Plasmodium falciparum DHFR-TS complexed with cycloguanil and NADPH | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2011-11-12 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Combined Spatial Limitation around Residues 16 and 108 of Plasmodium falciparum Dihydrofolate Reductase Explains Resistance to Cycloguanil.
Antimicrob.Agents Chemother., 56, 2012
|
|
3UR1
 
 | |
