5TXN
 
 | | STRUCTURE OF Q151M MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
3JA9
 
 | |
1AWC
 
 | | MOUSE GABP ALPHA/BETA DOMAIN BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*(BRU)P*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*AP*(CBR)P*AP*CP*(CBR)P*GP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*GP*(BRU)P*GP*(BRU)P*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*AP*T)-3'), PROTEIN (GA BINDING PROTEIN ALPHA), ... | | Authors: | Batchelor, A.H, Wolberger, C. | | Deposit date: | 1997-10-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of GABPalpha/beta: an ETS domain- ankyrin repeat heterodimer bound to DNA.
Science, 279, 1998
|
|
6JWP
 
 | | crystal structure of EGOC | | Descriptor: | Ego2, GTP-binding protein GTR1, GTP-binding protein GTR2, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2019-04-21 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the EGO-TC-mediated membrane tethering of the TORC1-regulatory Rag GTPases.
Sci Adv, 5, 2019
|
|
2WHX
 
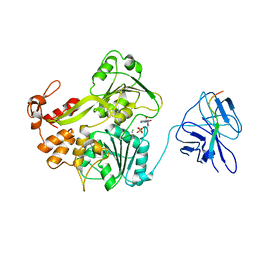 | | A second conformation of the NS3 protease-helicase from dengue virus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Luo, D, Lescar, J. | | Deposit date: | 2009-05-07 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexibility between the Protease and Helicase Domains of the Dengue Virus Ns3 Protein Conferred by the Linker Region and its Functional Implications.
J.Biol.Chem., 285, 2010
|
|
5VFU
 
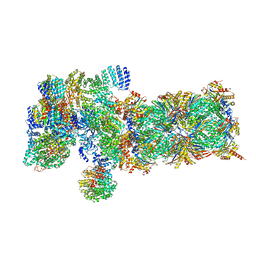 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
1P5O
 
 | |
8T2T
 
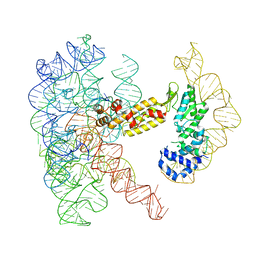 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | Descriptor: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
4M5V
 
 | | Influenza 2009 H1N1 endonuclease with 100 millimolar calcium | | Descriptor: | CALCIUM ION, Polymerase PA, SULFATE ION | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
6W4G
 
 | | Hepatitis C virus polymerase NS5B with RO inhibitor for SAR studies | | Descriptor: | 5-[[(2~{S})-4-[1,1-bis(oxidanylidene)-1,2-benzothiazol-3-yl]-2-~{tert}-butyl-3-oxidanyl-5-oxidanylidene-2~{H}-pyrrol-1-yl]methyl]-2-fluoranyl-benzenecarbonitrile, NS5B | | Authors: | Harris, S.F. | | Deposit date: | 2020-03-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of Noncompetitive Protein-Ligand Interactions for Structural Optimization.
J.Chem.Inf.Model., 60, 2020
|
|
4M5O
 
 | | 3-HYDROXY-6-PHENYL-1,2-DIHYDROPYRIDIN-2-ONE bound to influenza 2009 H1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-phenylpyridin-2(5H)-one, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
1P5P
 
 | |
2W2G
 
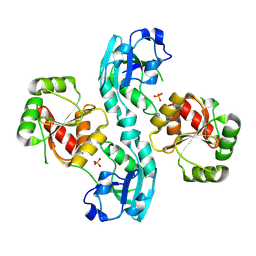 | | Human SARS coronavirus unique domain | | Descriptor: | NON-STRUCTURAL PROTEIN 3, SULFATE ION | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Kusov, Y, Hansen, G, Mesters, J.R, Schmidt, C.L, Hilgenfeld, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
7WJV
 
 | | Crystal structure of human liver FBPase complexed with an covalent inhibitor | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Cao, H, Huang, Y, Ren, Y, Wan, J. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | N -Acylamino Saccharin as an Emerging Cysteine-Directed Covalent Warhead and Its Application in the Identification of Novel FBPase Inhibitors toward Glucose Reduction.
J.Med.Chem., 65, 2022
|
|
4M5Q
 
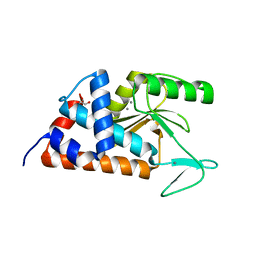 | | High-resolution apo influenza 2009 H1N1 endonuclease structure | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Polymerase PA | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
4M4Q
 
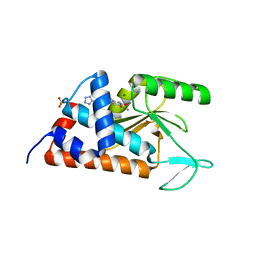 | | 6-(4-fluorophenyl)-3-hydroxy-5-[4-(1H-1,2,3,4-tetrazol-5-yl)phenyl] -1,2-dihydropyridin-2-one bound to influenza 2009 H1N1 endonuclease | | Descriptor: | 6-(4-fluorophenyl)-5-[4-(1H-tetrazol-5-yl)phenyl]pyridine-2,3-diol, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-08-07 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
1P5M
 
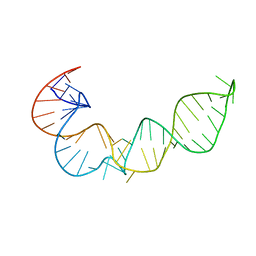 | |
6O3Y
 
 | | Crystal structure of yeast Nrd1 CID in complex with Sen1 NIM3 | | Descriptor: | CHLORIDE ION, Helicase SEN1, Protein NRD1 | | Authors: | Zhang, Y, Tong, L. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1.
Structure, 27, 2019
|
|
2AGQ
 
 | | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*CP*TP*AP*CP*AP*GP*GP*AP*CP*TP*(DOC))-3', 5'-D(*TP*CP*AP*TP*GP*AP*GP*TP*CP*CP*TP*GP*TP*AP*GP*CP*C)-3', ... | | Authors: | Ling, H, Yang, W. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis.
Embo J., 24, 2005
|
|
1AXC
 
 | | HUMAN PCNA | | Descriptor: | P21/WAF1, PCNA | | Authors: | Gulbis, J.M, Kuriyan, J. | | Deposit date: | 1997-10-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA.
Cell(Cambridge,Mass.), 87, 1996
|
|
1MIV
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme | | Descriptor: | MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
3TG5
 
 | | Structure of SMYD2 in complex with p53 and SAH | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
4M5R
 
 | | High-resolution influenza 2009 H1N1 endonuclease bound to 4-(1H-IMIDAZOL-1-YL)PHENOL | | Descriptor: | 1,2-ETHANEDIOL, 4-(1H-IMIDAZOL-1-YL)PHENOL, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
4JGH
 
 | | Structure of the SOCS2-Elongin BC complex bound to an N-terminal fragment of Cullin5 | | Descriptor: | Cullin-5, Suppressor of cytokine signaling 2, Transcription elongation factor B polypeptide 1, ... | | Authors: | Kim, Y.K, Kwak, M.J, Ku, B, Suh, H.Y, Joo, K, Lee, J, Jung, J.U, Oh, B.H. | | Deposit date: | 2013-03-01 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intersubunit recognition in elongin BC-cullin 5-SOCS box ubiquitin-protein ligase complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
