6MAF
 
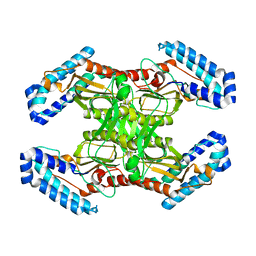 | | native BbvCI A2B2 tetramer at low resolution | | Descriptor: | BbvCI endonuclease subunit 1, BbvCI endonuclease subunit 2 | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Structure, subunit organization and behavior of the asymmetric Type IIT restriction endonuclease BbvCI.
Nucleic Acids Res., 47, 2019
|
|
3RPZ
 
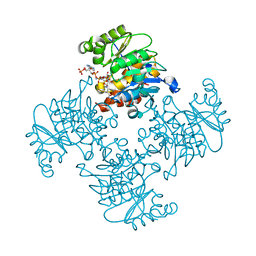 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with NADPH | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BETA-6-HYDROXY-1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
7AEU
 
 | |
7AET
 
 | |
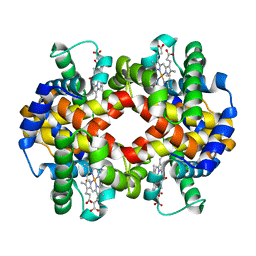
7AEV
 
 | |
6V2G
 
 | |
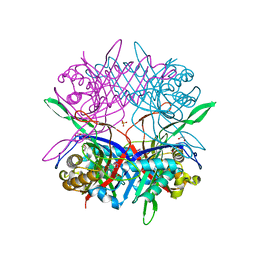
5Y2P
 
 | |
5Y9R
 
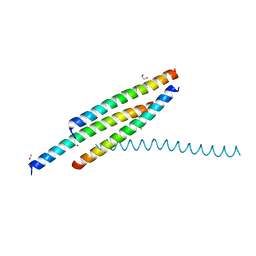 | |
8UUE
 
 | |
6V2C
 
 | |
5MEY
 
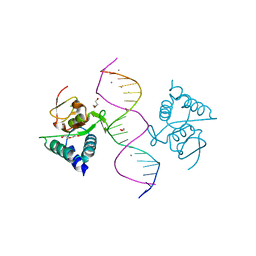 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
7AZ5
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
4R3U
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Mutase | | Descriptor: | 2-hydroxyisobutyryl-CoA mutase large subunit, 2-hydroxyisobutyryl-CoA mutase small subunit, 3-HYDROXYBUTANOYL-COENZYME A, ... | | Authors: | Zahn, M, Kurteva-Yaneva, N, Rohwerder, T, Straeter, N. | | Deposit date: | 2014-08-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the stereospecificity of bacterial B12-dependent 2-hydroxyisobutyryl-CoA mutase.
J.Biol.Chem., 290, 2015
|
|
3RTQ
 
 | | Structure of the mouse CD1d-HS44-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-05-03 | | Release date: | 2012-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional characterization of a novel nonglycosidic type I NKT agonist with immunomodulatory properties.
J.Immunol., 188, 2012
|
|
5LYN
 
 | | Structure of the Tpr Domain of Sgt2 in complex with yeast Ssa1 peptide fragment | | Descriptor: | PRO-THR-VAL-GLU-GLU-VAL-ASP, Small glutamine-rich tetratricopeptide repeat-containing protein 2, ZINC ION | | Authors: | Krysztofinska, E.M, Evans, N.J, Thapaliya, A, Murray, J.W, Morgan, R.M.L, Martinez-Lumbreras, S, Isaacson, R.L. | | Deposit date: | 2016-09-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Interactions of the TPR Domain of Sgt2 with Yeast Chaperones and Ybr137wp.
Front Mol Biosci, 4, 2017
|
|
4GTE
 
 | | T. Maritima FDTS (E144R mutant) with FAD and Folate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-({4-[(6aR)-3-amino-1-oxo-1,2,5,6,6a,7-hexahydroimidazo[1,5-f]pteridin-8(9H)-yl]phenyl}carbonyl)-L-glutamic acid, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A, Prabhakar, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GTA
 
 | | T. Maritima FDTS with FAD, dUMP, and Folinic Acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4E4H
 
 | | Crystal structure of Histone Demethylase NO66 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Wu, M, Tao, Y, Zang, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | tructural Basis for Tetramerization-dependent Gene Repression by Histone Demethylase NO66
To be Published
|
|
5F8A
 
 | | Crystal structure of the ternary EcoRV-DNA-Lu complex with uncleaved DNA substrate. Lanthanide binding to EcoRV-DNA complex inhibits cleavage. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*TP*CP*TP*TP*T)-3'), LUTETIUM (III) ION, ... | | Authors: | Sangani, S.S, Kehr, A.D, Sinha, K, Rule, G.S, Jen-Jacobson, L. | | Deposit date: | 2015-12-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Metal Ion Binding at the Catalytic Site Induces Widely Distributed Changes in a Sequence Specific Protein-DNA Complex.
Biochemistry, 55, 2016
|
|
5NM9
 
 | | Crystal structure of the placozoa Trichoplax adhaerens Smad4-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*AP*TP*GP*CP*GP*GP*GP*CP*GP*CP*GP*CP*CP*CP*GP*CP*AP*T)-3'), Mothers against decapentaplegic homolog, ZINC ION | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
3B04
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with oIPP. | | Descriptor: | 1-deoxy-1-[(4aR)-4a-[(2R)-1-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-3-methylidenepentan-2-yl]-7,8-dimethyl-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono-D-ribitol, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION | | Authors: | Unno, H, Nagai, T, Hemmi, H. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Covalent modification of reduced flavin mononucleotide in type-2 isopentenyl diphosphate isomerase by active-site-directed inhibitors.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5LU5
 
 | | A quantum half-site enzyme | | Descriptor: | 7-O-phosphono-D-glycero-alpha-D-manno-heptopyranose, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vivoli, M, Harmer, N.J, Pang, J. | | Deposit date: | 2016-09-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
5LTZ
 
 | | GmhA_mutant Q175E | | Descriptor: | 1,2-ETHANEDIOL, D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vivoli, M, Harmer, N.J. | | Deposit date: | 2016-09-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
4P3H
 
 | |
5MF0
 
 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
