7ZEU
 
 | |
7BFN
 
 | |
7BFU
 
 | |
7BFT
 
 | |
7BFO
 
 | |
7BFR
 
 | |
7BFV
 
 | | Thermogutta terrifontis esterase 2 phosphonylated by cyclosarin | | Descriptor: | Esterase, PHOSPHATE ION, SODIUM ION | | Authors: | Brazzolotto, X.B, Bzdrenga, J, Nachon, F. | | Deposit date: | 2021-01-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Thermophilic Bacterial Esterase for Scavenging Nerve Agents: A Kinetic, Biophysical and Structural Study.
Molecules, 26, 2021
|
|
6IJX
 
 | | Crystal Structure of AKR1C1 complexed with meclofenamic acid | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, L, Zhang, H, Chen, Y, Hu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
5GMI
 
 | | Crystal Structure of GRASP55 GRASP domain in complex with JAM-C C-terminus | | Descriptor: | Golgi reassembly-stacking protein 2, Junctional adhesion molecule C | | Authors: | Shi, N, Shi, X, Morelli, X, Betzi, S, Huang, X. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis
PLoS Genet., 13, 2017
|
|
2CQ7
 
 | | Solution structure of RSGI RUH-032, a cystein-rich domain of CRISP-2 from human cDNA | | Descriptor: | Cysteine-rich secretory protein-2 | | Authors: | Abe, T, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-032, a cystein-rich domain of CRISP-2 from human cDNA
To be Published
|
|
2D9P
 
 | | Solution structure of RNA binding domain 4 in Polyadenylation binding protein 3 | | Descriptor: | Polyadenylate-binding protein 3 | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-12 | | Release date: | 2006-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain 4 in Polyadenylation binding
protein 3
To be Published
|
|
5W58
 
 | | Crystal Complex of Cyclooxygenase-2: (S)-ARN-2508 (a dual COX and FAAH inhibitor) | | Descriptor: | (2S)-2-{2-fluoro-3'-[(hexylcarbamoyl)oxy][1,1'-biphenyl]-4-yl}propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Goodman, M.C, Banerjee, S, Piomelli, D, Marnett, L.J. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | Dual cyclooxygenase-fatty acid amide hydrolase inhibitor exploits novel binding interactions in the cyclooxygenase active site.
J. Biol. Chem., 293, 2018
|
|
3VU7
 
 | | Crystal structure of REV1-REV7-REV3 ternary complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Kikuchi, S, Hara, K, Shimizu, T, Sato, M, Hashimoto, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of recruitment of DNA polymerase [zeta] by interaction between REV1 and REV7 proteins
J.Biol.Chem., 287, 2012
|
|
3VQH
 
 | | Bromine SAD partially resolves multiple binding modes for PKA inhibitor H-89 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Pflug, A, Johnson, K.A, Engh, R.A. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anomalous dispersion analysis of inhibitor flexibility: a case study of the kinase inhibitor H-89
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7SL9
 
 | | CryoEM structure of SMCT1 | | Descriptor: | Sodium-coupled monocarboxylate transporter 1, butanoic acid, nanobody Nb2 | | Authors: | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | Deposit date: | 2021-10-23 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
3VAC
 
 | |
5WBG
 
 | | Crystal Structure of human Cytochrome P450 2B6 (Y226H/K262R) in complex with an analog of a drug Efavirenz | | Descriptor: | (2R,4S)-6-chloro-4-(cyclopropylethynyl)-2-methyl-4-(trifluoromethyl)-1,4-dihydro-2H-3,1-benzoxazine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, 5-cyclohexylpentan-1-ol, ... | | Authors: | Shah, M.B, Halpert, J.R. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog.
Int J Mol Sci, 19, 2018
|
|
7NAM
 
 | | LRP6_E1 in complex with Lr-EET-3.5 | | Descriptor: | Low-density lipoprotein receptor-related protein 6, SODIUM ION, Trypsin inhibitor 2, ... | | Authors: | Hansen, S, Hannoush, R.N. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution identifies high-affinity cystine-knot peptide agonists and antagonists of Wnt/ beta-catenin signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8JB7
 
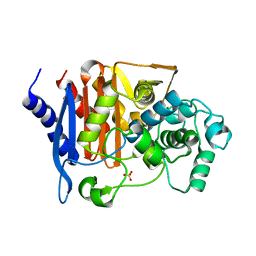 | | Crystal structure of CMY-185 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the molecular mechanism of high-level ceftazidime-avibactam resistance conferred by CMY-185.
Mbio, 15, 2024
|
|
8JB8
 
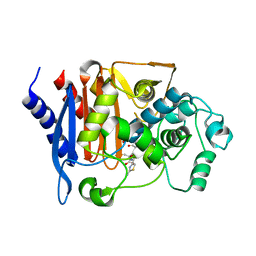 | | Crystal structure of CMY-185 complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the molecular mechanism of high-level ceftazidime-avibactam resistance conferred by CMY-185.
Mbio, 15, 2024
|
|
6CGC
 
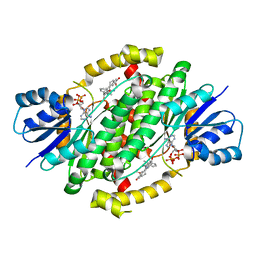 | | Crystal structure of human 17beta-HSD type 1 in ternary complex with 2-MeO-CC-156 and NADP+ | | Descriptor: | 3-{[(9beta,14beta,16alpha,17alpha)-3,17-dihydroxy-2-methoxyestra-1,3,5(10)-trien-16-yl]methyl}benzamide, Estradiol 17-beta-dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, T, Lin, S.X. | | Deposit date: | 2018-02-19 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Combined Biophysical Chemistry Reveals a New Covalent Inhibitor with a Low-Reactivity Alkyl Halide.
J Phys Chem Lett, 9, 2018
|
|
4F4X
 
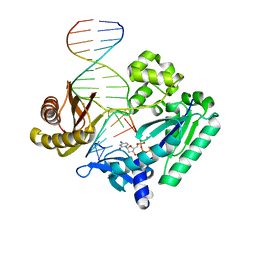 | | Y-family DNA polymerase chimera Dbh-Dpo4-Dpo4 #2 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*GP*C)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
4F4W
 
 | | Y-family DNA polymerase chimera Dbh-Dpo4-Dpo4 #1 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*C)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
4F4Y
 
 | | Y-family DNA polymerase chimera Dbh-Dpo4-Dbh | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*GP*CP*CP*C)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
4F4Z
 
 | | Y-family DNA polymerase chimera Dpo4-Dpo4-Dbh | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*GP*GP*GP*GP*GP*AP*AP*GP*CP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*CP*CP*CP*GP*GP*CP*TP*TP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
