3QXJ
 
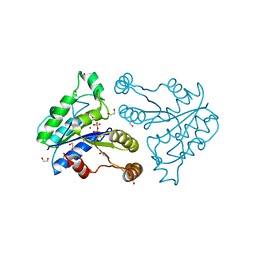 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GTP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QTP
 
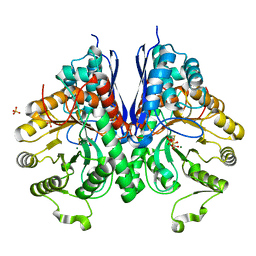 | |
3QY0
 
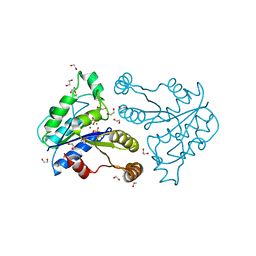 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
9DNU
 
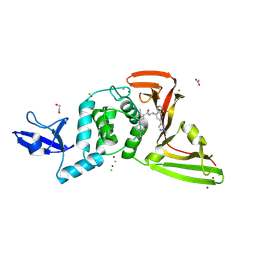 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun13296 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-5-[(1R,5S)-8-methyl-3,8-diazabicyclo[3.2.1]octan-3-yl]-N-{(1R)-1-[(2P)-2-(1-methyl-1H-pyrazol-4-yl)quinolin-4-yl]ethyl}benzamide, ACETATE ION, ... | | Authors: | Ansari, A, Jadhav, P, Arnold, E, Ruiz, F.X, Wang, J. | | Deposit date: | 2024-09-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of quinoline SARS-CoV-2 papain-like protease inhibitors as oral antiviral drug candidates.
Nat Commun, 16, 2025
|
|
5NYG
 
 | |
6LC5
 
 | | Crystal structure of barley exohydrolaseI W434F in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-4-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-oxane-2,3,5-triol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-11-17 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
3KWJ
 
 | | Structure of human DPP-IV with (2S,3S,11bS)-3-(3-Fluoromethyl-phenyl)-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-ylamine | | Descriptor: | (2S,3S,11bS)-3-[3-(fluoromethyl)phenyl]-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 soluble form | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Aryl- and heteroaryl-substituted aminobenzo[a]quinolizines as dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5ZFH
 
 | | Mouse Kallikrein 7 | | Descriptor: | Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2018-03-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-based drug design to overcome species differences in kallikrein 7 inhibition of 1,3,6-trisubstituted 1,4-diazepan-7-ones.
Bioorg. Med. Chem., 26, 2018
|
|
5T9J
 
 | | Crystal Structure of human GEN1 in complex with Holliday junction DNA in the upper interface | | Descriptor: | DNA (5'-D(*DAP*DCP*DGP*DAP*DTP*DGP*DGP*DAP*DGP*DCP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DCP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DAP*DTP*DTP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DAP*DGP*DGP*DGP*DAP*DTP*DGP*DC)-3'), DNA (5'-D(*DGP*DAP*DGP*DCP*DCP*DTP*DAP*DGP*DCP*DGP*DTP*DCP*DCP*DGP*DGP*DAP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Lee, S.-H, Biertumpfel, C. | | Deposit date: | 2016-09-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.00012732 Å) | | Cite: | Human Holliday junction resolvase GEN1 uses a chromodomain for efficient DNA recognition and cleavage.
Elife, 4, 2015
|
|
7FAR
 
 | | Crystal structure of PDE5A in complex with inhibitor L12 | | Descriptor: | 5-[bis(fluoranyl)methoxy]-2-[(4-chlorophenyl)methyl]-10-(3-methoxypropyl)-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.40006471 Å) | | Cite: | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
3R0F
 
 | | Human enterovirus 71 3C protease mutant H133G in complex with rupintrivir | | Descriptor: | 1,2-ETHANEDIOL, 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | Deposit date: | 2011-03-08 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3083 Å) | | Cite: | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
5EI5
 
 | | Crystal structure of MSF-aged Torpedo californica Acetylcholinesterase in complex with alkylene-linked bis-tacrine dimer (7 carbon linker) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Pesaresi, A, Lamba, D. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-09 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Disentangling the formation, mechanism, and evolvement of the covalent methanesulfonyl fluoride acetylcholinesterase adduct: Insights into an aged-like inactive complex susceptible to reactivation by a combination of nucleophiles.
Protein Sci., 33, 2024
|
|
3R1R
 
 | |
2Q73
 
 | | Crystal structure of iMazG from Vibrio DAT 722: Ctag-iMazG (P41212) | | Descriptor: | Hypothetical protein, MAGNESIUM ION | | Authors: | Robinson, A, Guilfoyle, A.P, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2007-06-05 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A putative house-cleaning enzyme encoded within an integron array: 1.8 A crystal structure defines a new MazG subtype.
Mol.Microbiol., 66, 2007
|
|
5OAX
 
 | | Galectin-3c in complex with thiogalactoside derivate | | Descriptor: | 5,6-bis(fluoranyl)-3-[[(2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-4-yl]oxymethyl]chromen-2-one, Galectin-3 | | Authors: | Nilsson, U.J, Peterson, K, Hakansson, M, Logan, D.T. | | Deposit date: | 2017-06-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Systematic Tuning of Fluoro-galectin-3 Interactions Provides Thiodigalactoside Derivatives with Single-Digit nM Affinity and High Selectivity.
J. Med. Chem., 61, 2018
|
|
7FAQ
 
 | | Crystal structure of PDE5A in complex with inhibitor L1 | | Descriptor: | 2-[(4-chlorophenyl)methyl]-5-methoxy-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.200136 Å) | | Cite: | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
3HWX
 
 | |
5DRR
 
 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-058 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
3HZP
 
 | |
3I0Y
 
 | |
2VFP
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V349L | | Descriptor: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
3AS3
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | Descriptor: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
4GQO
 
 | | 2.1 Angstrom resolution crystal structure of uncharacterized protein lmo0859 from Listeria monocytogenes EGD-e | | Descriptor: | Lmo0859 protein, TRIETHYLENE GLYCOL | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of uncharacterized protein lmo0859 from Listeria monocytogenes EGD-e
To be Published
|
|
1WCX
 
 | | Crystal Structure of Mutant Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (L75M/I193M/L248M, SeMet derivative, Form-1 crystal) | | Descriptor: | GLYCEROL, Uroporphyrinogen III Synthase | | Authors: | Mizohata, E, Matsuura, T, Murayama, K, Sakai, H, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-06 | | Release date: | 2005-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uroporphyrinogen III Synthase from Thermus thermophilus HB8
To be Published
|
|
3AUP
 
 | | Crystal structure of Basic 7S globulin from soybean | | Descriptor: | Basic 7S globulin | | Authors: | Yoshizawa, T, Shimizu, T, Taichi, M, Nishiuchi, Y, Yamabe, M, Shichijo, N, Unzai, S, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of basic 7S globulin, a xyloglucan-specific endo-beta-1,4-glucanase inhibitor protein-like protein from soybean lacking inhibitory activity against endo-beta-glucanase
Febs J., 278, 2011
|
|
