5HHJ
 
 | |
5IRG
 
 | |
5IRF
 
 | |
8BGJ
 
 | |
3FQD
 
 | | Crystal Structure of the S. pombe Rat1-Rai1 Complex | | Descriptor: | 5'-3' exoribonuclease 2, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2009-01-07 | | Release date: | 2009-02-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1.
Nature, 458, 2009
|
|
4GUA
 
 | | Alphavirus P23pro-zbd | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural polyprotein, SULFATE ION, ... | | Authors: | Shin, G, Yost, S, Miller, M, Marcotrigiano, J. | | Deposit date: | 2012-08-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structural and functional insights into alphavirus polyprotein processing and pathogenesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5HHL
 
 | |
5BUH
 
 | | Influenza PB2 bound to a hydroxymethyl azaindole inhibitor | | Descriptor: | N-[(1R,3S)-3-({5-fluoro-2-[5-fluoro-2-(hydroxymethyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]pyrimidin-4-yl}amino)cyclohexyl]pyrrolidine-1-carboxamide, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-06-03 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Novel 2-Substituted 7-Azaindole and 7-Azaindazole Analogues as Potential Antiviral Agents for the Treatment of Influenza.
ACS Med Chem Lett, 8, 2017
|
|
5HHK
 
 | |
5W6Z
 
 | | Crystal structure of the H24W mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5W6X
 
 | |
3FQG
 
 | | Crystal Structure of the S. pombe Rai1 | | Descriptor: | MAGNESIUM ION, Protein din1 | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2009-01-07 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1.
Nature, 458, 2009
|
|
7PY2
 
 | | Structure of pathological TDP-43 filaments from ALS with FTLD | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Arseni, D, Hasegawa, H, Murzin, A.G, Kametani, F, Arai, M, Yoshida, M, Falcon, B. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure of pathological TDP-43 filaments from ALS with FTLD.
Nature, 601, 2022
|
|
5VY2
 
 | | Crystal structure of the F36A mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-05-24 | | Release date: | 2018-11-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5WJI
 
 | | Crystal structure of the F61S mutant of HsNUDT16 | | Descriptor: | ACETIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-07-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
8AVG
 
 | | Cryo-EM structure of mouse Elp123 with bound SAM | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
5LCJ
 
 | | In-Gel Activity-Based Protein Profiling of a Clickable Covalent Erk 1/2 Inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [(1~{R},4~{Z})-cyclooct-4-en-1-yl] ~{N}-[4-[4-[[5-chloranyl-4-[[2-(propanoylamino)phenyl]amino]pyrimidin-2-yl]amino]pyridin-2-yl]but-3-ynyl]carbamate | | Authors: | O'Reilly, M, Wright, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-20 | | Last modified: | 2016-08-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | In-gel activity-based protein profiling of a clickable covalent ERK1/2 inhibitor.
Mol Biosyst, 12, 2016
|
|
5LCK
 
 | | A Clickable Covalent ERK 1/2 Inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, ~{N}-[2-[[2-[(5-methoxypyridin-3-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]propanamide | | Authors: | O'Reilly, M, Wright, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-20 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In-gel activity-based protein profiling of a clickable covalent ERK1/2 inhibitor.
Mol Biosyst, 12, 2016
|
|
1VYM
 
 | | NATIVE HUMAN PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M. | | Deposit date: | 2004-05-03 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1VYJ
 
 | | Structural and biochemical studies of human PCNA complexes provide the basis for association with CDK/cyclin and rationale for inhibitor design | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN, SMALL PEPTIDE SAVLQKKITDYFHPKK | | Authors: | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M.D. | | Deposit date: | 2004-04-30 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1W60
 
 | | NATIVE HUMAN PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M. | | Deposit date: | 2004-08-11 | | Release date: | 2005-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7W1M
 
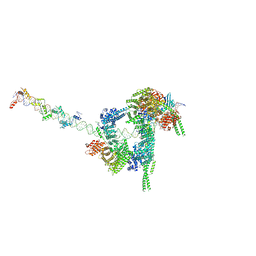 | | Cryo-EM structure of human cohesin-CTCF-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cohesin subunit SA-1, ... | | Authors: | Shi, Z.B, Bai, X.C, Yu, H. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | CTCF and R-loops are boundaries of cohesin-mediated DNA looping.
Mol.Cell, 83, 2023
|
|
7F5S
 
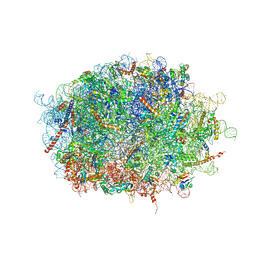 | | human delta-METTL18 60S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Takahashi, M, Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance.
Elife, 11, 2022
|
|
4S0H
 
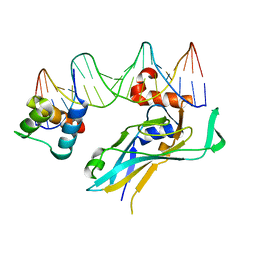 | | TBX5 DB, NKX2.5 HD, ANF DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*TP*TP*CP*AP*AP*AP*GP*GP*TP*GP*TP*GP*AP*GP*A)-3', 5'-D(*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP*TP*GP*AP*AP*GP*TP*GP*G)-3', Homeobox protein Nkx-2.5, ... | | Authors: | Pradhan, L. | | Deposit date: | 2014-12-31 | | Release date: | 2015-12-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5.
Biochemistry, 55, 2016
|
|
7FJT
 
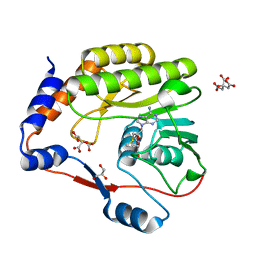 | |
