4RZR
 
 | | Bypass of a bulky adduct dG1,8 by DPO4 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Vyas, R, Suo, Z. | | Deposit date: | 2014-12-23 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Basis for the Bypass of a Bulky DNA Adduct Catalyzed by a Y-Family DNA Polymerase.
J.Am.Chem.Soc., 137, 2015
|
|
1CF4
 
 | | CDC42/ACK GTPASE-BINDING DOMAIN COMPLEX | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (ACTIVATED P21CDC42HS KINASE), ... | | Authors: | Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Lim, L, Laue, E.D. | | Deposit date: | 1999-03-23 | | Release date: | 1999-06-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK.
Nature, 399, 1999
|
|
1GRN
 
 | | CRYSTAL STRUCTURE OF THE CDC42/CDC42GAP/ALF3 COMPLEX. | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nassar, N, Hoffman, G.R, Clardy, J.C, Cerione, R.A. | | Deposit date: | 1998-07-30 | | Release date: | 1999-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP.
Nat.Struct.Biol., 5, 1998
|
|
1GZS
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN THE GEF DOMAIN OF THE SALMONELLA TYPHIMURIUM SOPE TOXIN AND HUMAN Cdc42 | | Descriptor: | GTP-BINDING PROTEIN, SOPE, SULFATE ION | | Authors: | Buchwald, G, Friebel, A, Galan, J.E, Hardt, W.D, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 2002-06-05 | | Release date: | 2002-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Reversible Activation of a Rho Protein by the Bacterial Toxin Sope
Embo J., 21, 2002
|
|
1GUA
 
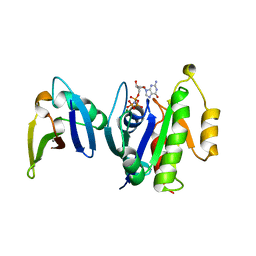 | | HUMAN RAP1A, RESIDUES 1-167, DOUBLE MUTANT (E30D,K31E) COMPLEXED WITH GPPNHP AND THE RAS-BINDING-DOMAIN OF HUMAN C-RAF1, RESIDUES 51-131 | | Descriptor: | C-RAF1, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Nassar, N, Wittinghofer, A. | | Deposit date: | 1996-06-18 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ras/Rap effector specificity determined by charge reversal.
Nat.Struct.Biol., 3, 1996
|
|
5UYK
 
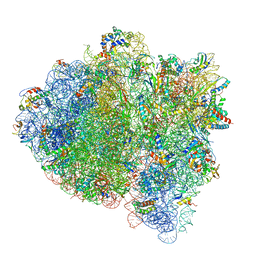 | | 70S ribosome bound with cognate ternary complex not base-paired to A site codon (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
1MAH
 
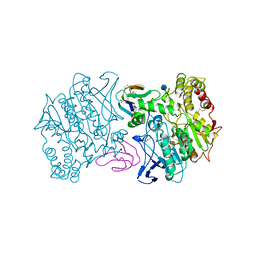 | | FASCICULIN2-MOUSE ACETYLCHOLINESTERASE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, FASCICULIN 2 | | Authors: | Bourne, Y, Taylor, P, Marchot, P. | | Deposit date: | 1995-11-21 | | Release date: | 1996-04-03 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Acetylcholinesterase inhibition by fasciculin: crystal structure of the complex.
Cell(Cambridge,Mass.), 83, 1995
|
|
5V9X
 
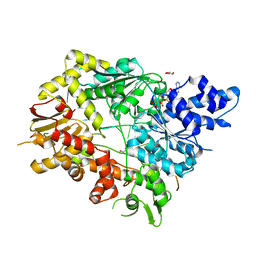 | | Structure of Mycobacterium smegmatis helicase Lhr bound to ssDNA and AMP-PNP | | Descriptor: | ATP-dependent DNA helicase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ordonez, H, Jacewicz, A, Ferrao, R, Shuman, S. | | Deposit date: | 2017-03-23 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structure of mycobacterial 3'-to-5' RNA:DNA helicase Lhr bound to a ssDNA tracking strand highlights distinctive features of a novel family of bacterial helicases.
Nucleic Acids Res., 46, 2018
|
|
6QVJ
 
 | | HsCKK (human CAMSAP1) decorated 14pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
1LXG
 
 | |
6QNO
 
 | | Rhodopsin-Gi protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab antibody fragment heavy chain, Fab antibody fragment light chain, ... | | Authors: | Tsai, C.-J, Marino, J, Adaixo, R.J, Pamula, F, Muehle, J, Maeda, S, Flock, T, Taylor, N.M.I, Mohammed, I, Matile, H, Dawson, R.J.P, Deupi, X, Stahlberg, H, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
5VDI
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Mutant N38Q Bound to AM-3607, Glycine, and Ivermectin | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Human GlyR alpha 3 Bound to Ivermectin.
Structure, 25, 2017
|
|
1MFQ
 
 | | Crystal Structure Analysis of a Ternary S-Domain Complex of Human Signal Recognition Particle | | Descriptor: | 7S RNA of human SRP, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kuglstatter, A, Oubridge, C, Nagai, K. | | Deposit date: | 2002-08-13 | | Release date: | 2002-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Induced structural changes of 7SL RNA during the assembly of human
signal recognition particle
Nat.Struct.Biol., 9, 2002
|
|
5GYN
 
 | |
1MGN
 
 | |
5VHB
 
 | |
5VIB
 
 | |
5GJE
 
 | | Three-dimensional reconstruction of human LRP6 ectodomain complexed with Dkk1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matoba, K, Mihara, E, Tamura-Kawakami, K, Hirai, H, Thompson, S, Iwasaki, K, Takagi, J. | | Deposit date: | 2016-06-29 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Conformational Freedom of the LRP6 Ectodomain Is Regulated by N-glycosylation and the Binding of the Wnt Antagonist Dkk1
Cell Rep, 18, 2017
|
|
1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
6QWS
 
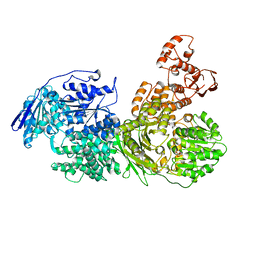 | |
1MCO
 
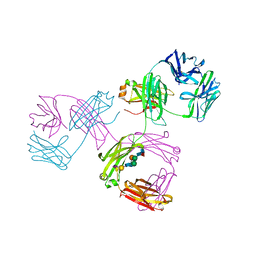 | | THREE-DIMENSIONAL STRUCTURE OF A HUMAN IMMUNOGLOBULIN WITH A HINGE DELETION | | Descriptor: | IGG1 MCG INTACT ANTIBODY (HEAVY CHAIN), IGG1 MCG INTACT ANTIBODY (LIGHT CHAIN), N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-L-gulopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guddat, L.W, Edmundson, A.B. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Three-dimensional structure of a human immunoglobulin with a hinge deletion.
Proc.Natl.Acad.Sci.Usa, 90, 1993
|
|
5W0P
 
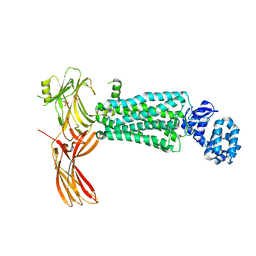 | | Crystal structure of rhodopsin bound to visual arrestin determined by X-ray free electron laser | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, He, Y, de Waal, P.W, Gao, X, Kang, Y, Van Eps, N, Yin, Y, Pal, K, Goswami, D, White, T.A, Barty, A, Latorraca, N.R, Chapman, H.N, Hubbell, W.L, Dror, R.O, Stevens, R.C, Cherezov, V, Gurevich, V.V, Griffin, P.R, Ernst, O.P, Melcher, K, Xu, H.E. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors.
Cell, 170, 2017
|
|
5GW0
 
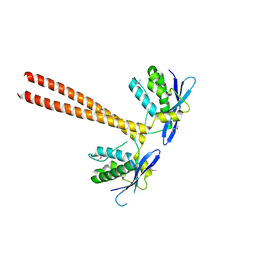 | | Crystal structure of SNX16 PX-Coiled coil | | Descriptor: | Sorting nexin-16 | | Authors: | Xu, J, Liu, J. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | SNX16 Regulates the Recycling of E-Cadherin through a Unique Mechanism of Coordinated Membrane and Cargo Binding.
Structure, 25, 2017
|
|
1MP8
 
 | | Crystal structure of Focal Adhesion Kinase (FAK) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, focal adhesion kinase 1 | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C.G, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography
Structure, 10, 2002
|
|
5W34
 
 | |
