4AC7
 
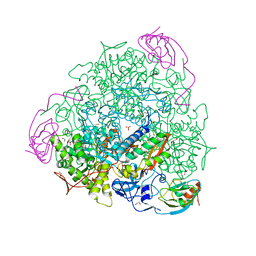 | | The crystal structure of Sporosarcina pasteurii urease in complex with citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, HYDROXIDE ION, ... | | Authors: | Benini, S, Kosikowska, P, Cianci, M, Gonzalez Vara, A, Berlicki, L, Ciurli, S. | | Deposit date: | 2011-12-14 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of Sporosarcina Pasteurii Urease in a Complex with Citrate Provides New Hints for Inhibitor Design.
J.Biol.Inorg.Chem., 18, 2013
|
|
1OIT
 
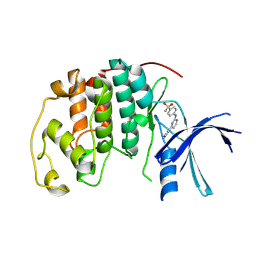 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | 4-[(4-IMIDAZO[1,2-A]PYRIDIN-3-YLPYRIMIDIN-2-YL)AMINO]BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
2RIQ
 
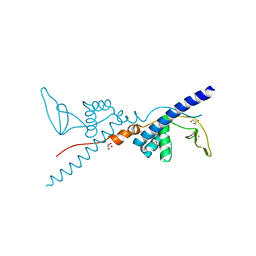 | | Crystal Structure of the Third Zinc-binding domain of human PARP-1 | | Descriptor: | ETHANOL, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.F, Servent, K.M. | | Deposit date: | 2007-10-12 | | Release date: | 2008-01-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Third Zinc-binding Domain of Human Poly(ADP-ribose) Polymerase-1 Coordinates DNA-dependent Enzyme Activation.
J.Biol.Chem., 283, 2008
|
|
3E02
 
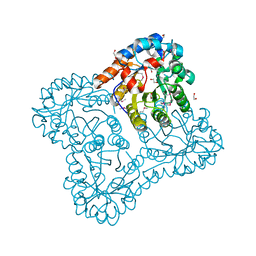 | |
3S5Q
 
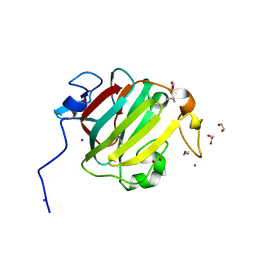 | |
3DPK
 
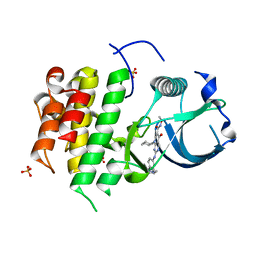 | | cFMS tyrosine kinase in complex with a pyridopyrimidinone inhibitor | | Descriptor: | 8-cyclohexyl-N-methoxy-5-oxo-2-{[4-(2-pyrrolidin-1-ylethyl)phenyl]amino}-5,8-dihydropyrido[2,3-d]pyrimidine-6-carboxamide, Macrophage colony-stimulating factor 1 receptor, Fibroblast growth factor receptor 1, ... | | Authors: | Schubert, C. | | Deposit date: | 2008-07-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrido[2,3-d]pyrimidin-5-ones: a novel class of antiinflammatory macrophage colony-stimulating factor-1 receptor inhibitors
J.Med.Chem., 52, 2009
|
|
2RUS
 
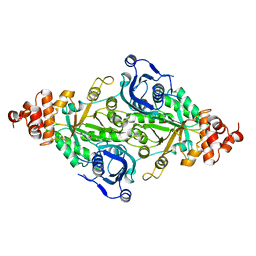 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE, MG(II), AND ACTIVATOR CO2 AT 2.3-ANGSTROMS RESOLUTION | | Descriptor: | FORMYL GROUP, MAGNESIUM ION, RUBISCO (RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE) | | Authors: | Lundqvist, T, Schneider, G. | | Deposit date: | 1991-10-11 | | Release date: | 1991-10-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ternary complex of ribulose-1,5-bisphosphate carboxylase, Mg(II), and activator CO2 at 2.3-A resolution.
Biochemistry, 30, 1991
|
|
4MC6
 
 | | HIV protease in complex with SA499 | | Descriptor: | 1,2-ETHANEDIOL, 1-tert-butyl-3-{(2S,3R)-4-[(4R)-7-fluoro-1,1-dioxido-4-(propan-2-yl)-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}urea, CHLORIDE ION, ... | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
4H0A
 
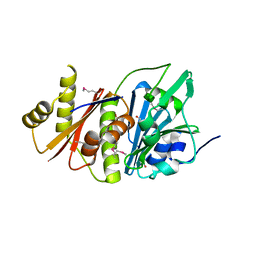 | |
2IEI
 
 | | Crystal structure of rabbit muscle glycogen phosphorylase in complex with 3,4-dihydro-2-quinolone | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, (S)-2-CHLORO-N-(1-(2-(2-HYDROXYETHYLAMINO)-2-OXOETHYL)-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-3-YL)-6H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, Glycogen phosphorylase, ... | | Authors: | Birch, A.M, Kenny, P.W, Oikonomakos, N.G, Otterbein, L, Schofield, P, Whittamore, P.R.O, Whalley, D.P, Rowsell, S, Pauptit, R, Pannifer, A, Breed, J, Minshull, C. | | Deposit date: | 2006-09-19 | | Release date: | 2006-12-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Development of potent, orally active 1-substituted-3,4-dihydro-2-quinolone glycogen phosphorylase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4OBC
 
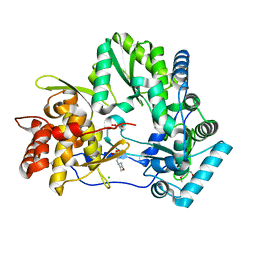 | | Crystal structure of HCV polymerase NS5b genotype 2a JFH-1 isolate with the S15G, C223H, V321I resistance mutations against the guanosine analog GS-0938 (PSI-3529238) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Edwards, T.E, Abendroth, J, Appleby, T.C. | | Deposit date: | 2014-01-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular and Structural Basis for the Roles of Hepatitis C Virus Polymerase NS5B Amino Acids 15, 223, and 321 in Viral Replication and Drug Resistance.
Antimicrob.Agents Chemother., 58, 2014
|
|
1AGC
 
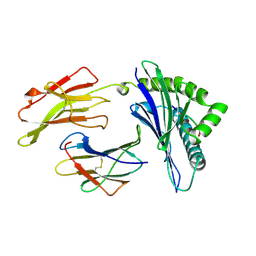 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYQL-7Q MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYQL - 7Q MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
3TYT
 
 | |
2ISW
 
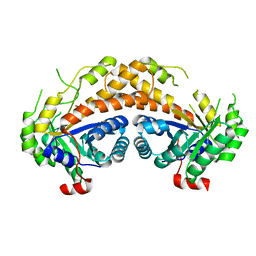 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2006-10-18 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia.
J.Biol.Chem., 282, 2007
|
|
4R3B
 
 | |
5SYZ
 
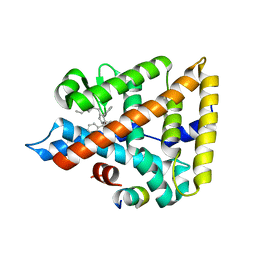 | | Human Liver Receptor Homologue-1 (LRH-1) Bound to a RJW100 stereoisomer and a Fragment of TIF-2 | | Descriptor: | (1S,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-ol, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Ortlund, E.A, Mays, S.G. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9261 Å) | | Cite: | Crystal Structures of the Nuclear Receptor, Liver Receptor Homolog 1, Bound to Synthetic Agonists.
J. Biol. Chem., 291, 2016
|
|
3SWA
 
 | | E. Cloacae MurA R120A complex with UNAG and covalent adduct of PEP with CYS115 | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Han, H, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
4J37
 
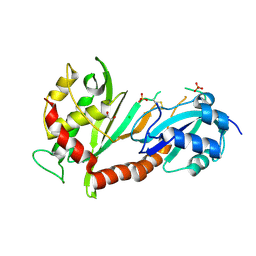 | | Crystal structure of the catalytic domain of human Pus1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In Human Pseudouridine Synthase 1 (hPus1), a C-Terminal Helical Insert Blocks tRNA from Binding in the Same Orientation as in the Pus1 Bacterial Homologue TruA, Consistent with Their Different Target Selectivities.
J.Mol.Biol., 425, 2013
|
|
3CTQ
 
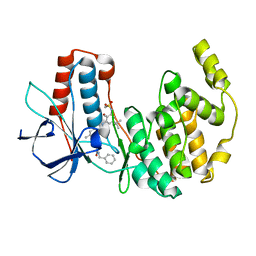 | | Structure of MAP kinase p38 in complex with a 1-o-tolyl-1,2,3-triazole-4-carboxamide | | Descriptor: | Mitogen-activated protein kinase 14, N-benzyl-1-[5-({5-tert-butyl-2-methoxy-3-[(methylsulfonyl)amino]phenyl}carbamoyl)-2-methylphenyl]-1H-1,2,3-triazole-4-carboxamide | | Authors: | Qian, K. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design and subsequent optimization of 2-tolyl-(1,2,3-triazol-1-yl-4-carboxamide) inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3FXA
 
 | |
3G8Y
 
 | |
3CY2
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand II | | Descriptor: | (4R)-7-chloro-9-methyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Huber, K, Bracher, F, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 7,8-Dichloro-1-oxo-beta-carbolines as a Versatile Scaffold for the Development of Potent and Selective Kinase Inhibitors with Unusual Binding Modes
J.Med.Chem., 55, 2012
|
|
1NUW
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate and Phosphate at pH 9.6 | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-01 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metaphosphate in the active site of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
4AD3
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with Glucose-1,3-deoxymannojirimycin | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1NUX
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate and inhibitory concentrations of Potassium (200mM) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-01 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metaphosphate in the active site of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
