3WIY
 
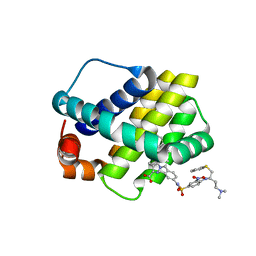 | | Crystal structure of Mcl-1 in complex with compound 10 | | Descriptor: | 7-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]carbamoyl}-2-methylphenyl)-3-[3-(naphthalen-1-yloxy)propyl]pyrazolo[1,5-a]pyridine-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Sogabe, S, Igaki, S, Hayano, Y. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
J.Med.Chem., 56, 2013
|
|
3G3E
 
 | |
3WYM
 
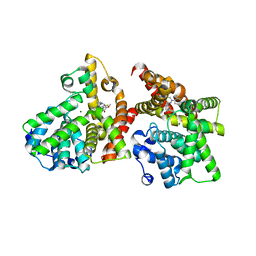 | | Crystal structure of the catalytic domain of PDE10A complexed with 1-(2-fluoro-4-(1H-pyrazol-1-yl)phenyl)-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one | | Descriptor: | 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Hayano, Y. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one (TAK-063), a highly potent, selective, and orally active phosphodiesterase 10A (PDE10A) inhibitor.
J.Med.Chem., 57, 2014
|
|
5AXP
 
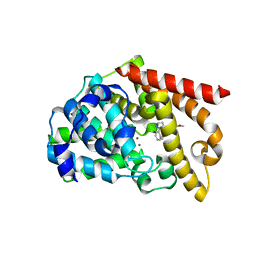 | | Crystal structure of the catalytic domain of PDE10A complexed with 1-(2-fluoro-4-(2-oxo-1,3-oxazolidin-3-yl)phenyl)-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one | | Descriptor: | 3-[3-fluoranyl-4-[5-methoxy-4-oxidanylidene-3-(2-phenylpyrazol-3-yl)pyridazin-1-yl]phenyl]-1,3-oxazolidin-2-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2015-07-31 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and synthesis of a novel 2-oxindole scaffold as a highly potent and brain-penetrant phosphodiesterase 10A inhibitor
Bioorg.Med.Chem., 23, 2015
|
|
2BDY
 
 | | thrombin in complex with inhibitor | | Descriptor: | Hirudin IIIB', N-(4-CARBAMIMIDOYL-BENZYL)-2-[2-HYDROXY-6-METHYL-3-(NAPHTHALENE-1-SULFONYLAMINO)-PHENYL]-ACETAMIDE, SODIUM ION, ... | | Authors: | Xue, Y. | | Deposit date: | 2005-10-21 | | Release date: | 2006-10-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Phenolic P2/P3 core motif as thrombin inhibitors--design, synthesis, and X-ray co-crystal structure.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4MUW
 
 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 2-{4-[(6,7-difluoro-1H-benzimidazol-2-yl)amino]phenoxy}-N-methyl-3,4'-bipyridin-2'-amine, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
3VF8
 
 | |
3WYK
 
 | | Crystal structure of the catalytic domain of PDE10A complexed with 3-(1-phenyl-1H-pyrazol-5-yl)-1-(3-(trifluoromethyl)phenyl)pyridazin-4(1H)-one | | Descriptor: | 3-(1-phenyl-1H-pyrazol-5-yl)-1-[3-(trifluoromethyl)phenyl]pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Hayano, Y. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one (TAK-063), a highly potent, selective, and orally active phosphodiesterase 10A (PDE10A) inhibitor.
J.Med.Chem., 57, 2014
|
|
3WYL
 
 | | Crystal structure of the catalytic domain of PDE10A complexed with 5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)-1-(3-(trifluoromethyl)phenyl)pyridazin-4(1H)-one | | Descriptor: | 5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)-1-[3-(trifluoromethyl)phenyl]pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Hayano, Y. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one (TAK-063), a highly potent, selective, and orally active phosphodiesterase 10A (PDE10A) inhibitor.
J.Med.Chem., 57, 2014
|
|
4CSS
 
 | | Crystal structure of FimH in complex with a sulfonamide biphenyl alpha D-mannoside | | Descriptor: | 4'-(alpha-D-Mannopyranosyloxy)-biphenyl-4-methyl sulfonamide, GLYCEROL, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
3VF9
 
 | |
3S9S
 
 | | Ligand binding domain of PPARgamma complexed with a benzimidazole partial agonist | | Descriptor: | 1-(3,4-dichlorobenzyl)-2-methyl-N-[(1R)-1-phenylpropyl]-1H-benzimidazole-5-carboxamide, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Lambert, M.H, Xu, R.X. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of GSK1997132B a novel centrally penetrant benzimidazole PPARgamma partial agonist.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3WIX
 
 | | Crystal structure of Mcl-1 in complex with compound 4 | | Descriptor: | 7-(4-carboxyphenyl)-3-[3-(naphthalen-1-yloxy)propyl]pyrazolo[1,5-a]pyridine-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Sogabe, S, Igaki, S, Hayano, Y. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
J.Med.Chem., 56, 2013
|
|
3TU7
 
 | |
4J70
 
 | | Yeast 20S proteasome in complex with the belactosin derivative 3e | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Kawamura, S, Unno, Y, List, A, Tanaka, M, Sasaki, T, Arisawa, M, Asai, A, Groll, M, Shuto, S. | | Deposit date: | 2013-02-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent Proteasome Inhibitors Derived from the Unnatural cis-Cyclopropane Isomer of Belactosin A: Synthesis, Biological Activity, and Mode of Action.
J.Med.Chem., 56, 2013
|
|
3MSJ
 
 | | Structure of bace (beta secretase) in complex with inhibitor | | Descriptor: | 3-(2-amino-5-chloro-1H-benzimidazol-1-yl)propan-1-ol, BETA-SECRETASE 1, GLYCEROL | | Authors: | Madden, J, Kramer, J, Smith, M.A, Barker, J, Godemann, R. | | Deposit date: | 2010-04-29 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2R6N
 
 | | Crystal structure of a pyrrolopyrimidine inhibitor in complex with human Cathepsin K | | Descriptor: | 1-{7-cyclohexyl-6-[4-(4-methylpiperazin-1-yl)benzyl]-7H-pyrrolo[2,3-d]pyrimidin-2-yl}methanamine, Cathepsin K | | Authors: | Cowan-Jacob, S.W, Ramage, P, Mathis, B, Geisse, S. | | Deposit date: | 2007-09-06 | | Release date: | 2007-11-06 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel scaffold for cathepsin K inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5SU5
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03E05 from the F2X-Universal Library | | Descriptor: | 5-bromo-1-methylpyrimidine-2,4(1H,3H)-dione, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
4FSE
 
 | |
5STN
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03A11 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-methyl-1-{(1S,2R)-2-[(1,2,4-triazolidin-1-yl)methyl]cyclohexyl}methanamine, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
3VW8
 
 | | Crystal structure of human c-Met kinase domain with its inhibitor | | Descriptor: | CHLORIDE ION, Hepatocyte growth factor receptor, N-({4-[(6,7-dimethoxyquinolin-4-yl)oxy]phenyl}carbamothioyl)-2-phenylacetamide | | Authors: | Matsumoto, S, Miyamoto, N, Hirayama, T, Oki, H, Okada, K, Tawada, M, Iwata, H, Miki, H, Nakamura, K, Hori, A, Imamura, S. | | Deposit date: | 2012-08-08 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design, synthesis, and evaluation of imidazo[1,2-b]pyridazine and imidazo[1,2-a]pyridine derivatives as novel dual c-Met and VEGFR2 kinase inhibitors.
Bioorg.Med.Chem., 21, 2013
|
|
5SU9
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03F06 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N~2~-methyl-N~2~-[2-methyl-2-(4-methylphenyl)propyl]glycinamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
5SUF
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03H03 from the F2X-Universal Library | | Descriptor: | 1-benzyl-1H-pyrazole-4-carboxylic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
5SUC
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03G02 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-(2-hydroxyethyl)-N-methyl-2-phenylacetamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
5STB
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P02F05 from the F2X-Universal Library | | Descriptor: | 3-ethyl-5-(ethylsulfanyl)-4H-1,2,4-triazol-4-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
