1NJS
 
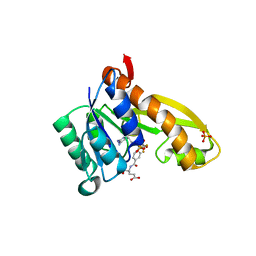 | | human GAR Tfase in complex with hydrolyzed form of 10-trifluoroacetyl-5,10-dideaza-acyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Marsilje, T.H, Li, C, Hedrick, M.P, Gooljarsingh, L.T, Tavassoli, A, Benkovic, S.J, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational Design, Synthesis, Evaluation, and Crystal Structure of a Potent Inhibitor of Human GAR Tfase: 10-(Trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic Acid
Biochemistry, 42, 2003
|
|
2N0I
 
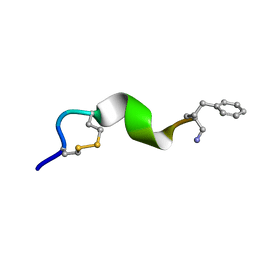 | | NMR solution structure for di-sulfide 11mer peptide | | Descriptor: | di-sulfide 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2MV8
 
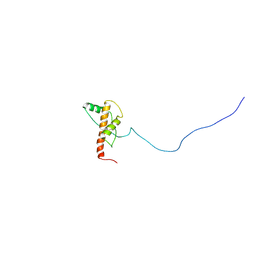 | | Solution structure of Ovis Aries PrP with mutation delta190-197 | | Descriptor: | Major prion protein | | Authors: | Munoz, C, Egalon, A, Beringue, V, Rezaei, H, Dron, M, Sizun, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Generating Bona Fide Mammalian Prions with Internal Deletions.
J.Virol., 90, 2016
|
|
2N4N
 
 | |
7WIZ
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, GLUTAMINE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liu, J.L, Guo, C.J. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7WJ4
 
 | |
1I5E
 
 | |
2MV9
 
 | | Solution structure of Ovis Aries PrP with mutation delta193-196 | | Descriptor: | Major prion protein | | Authors: | Munoz, C, Egalon, A, Beringue, V, Rezaei, H, Dron, M, Sizun, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Generating Bona Fide Mammalian Prions with Internal Deletions.
J.Virol., 90, 2016
|
|
2XNS
 
 | | Crystal Structure Of Human G alpha i1 Bound To A Designed Helical Peptide Derived From The Goloco Motif Of RGS14 | | Descriptor: | GUANINE NUCLEOTIDE-BINDING PROTEIN G(I) SUBUNIT ALPHA-1, GUANOSINE-5'-DIPHOSPHATE, REGULATOR OF G-PROTEIN SIGNALING 14, ... | | Authors: | Bosch, D, Sammond, D.W, Butterfoss, G.L, Machius, M, Siderovski, D.P, Kuhlman, B. | | Deposit date: | 2010-08-05 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Computational Design of the Sequence and Structure of a Protein-Binding Peptide.
J.Am.Chem.Soc., 133, 2011
|
|
7WKC
 
 | |
1KA9
 
 | |
1K46
 
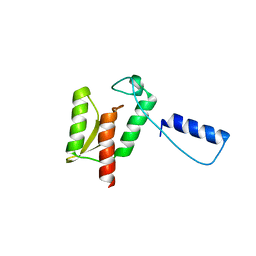 | | Crystal Structure of the Type III Secretory Domain of Yersinia YopH Reveals a Domain-Swapped Dimer | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH | | Authors: | Smith, C.L, Khandelwal, P, Keliikuli, K, Zuiderweg, E.R.P, Saper, M.A. | | Deposit date: | 2001-10-05 | | Release date: | 2001-11-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the type III secretion and substrate-binding domain of Yersinia YopH phosphatase.
Mol.Microbiol., 42, 2001
|
|
2N7T
 
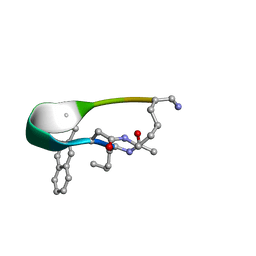 | | NMR structure of Peptide PG-992 in DPC micelles | | Descriptor: | Peptide PG-992 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
2N7N
 
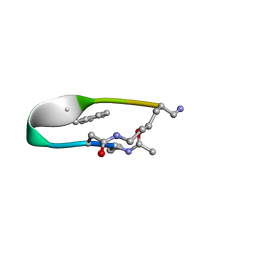 | | NMR structure of Peptide PG-989 in DPC micelles | | Descriptor: | Peptide PG-989 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
3STT
 
 | |
2N6I
 
 | |
2NBL
 
 | | Peptide model of 4-stranded beta-arch | | Descriptor: | Designed beta-arch | | Authors: | Kung, V.M. | | Deposit date: | 2016-03-05 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | A designed peptide model of early-stage amyloid structures
To be Published
|
|
1PEF
 
 | |
1PMN
 
 | | Crystal structure of JNK3 in complex with an imidazole-pyrimidine inhibitor | | Descriptor: | CYCLOPROPYL-{4-[5-(3,4-DICHLOROPHENYL)-2-[(1-METHYL)-PIPERIDIN]-4-YL-3-PROPYL-3H-IMIDAZOL-4-YL]-PYRIMIDIN-2-YL}AMINE, Mitogen-activated protein kinase 10 | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | Deposit date: | 2003-06-11 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
1JR1
 
 | | Crystal structure of Inosine Monophosphate Dehydrogenase in complex with Mycophenolic Acid | | Descriptor: | INOSINIC ACID, Inosine-5'-Monophosphate Dehydrogenase 2, MYCOPHENOLIC ACID, ... | | Authors: | Sintchak, M.D, Fleming, M.A, Futer, O, Raybuck, S.A, Chambers, S.P, Caron, P.R, Murcko, M.A, Wilson, K.P. | | Deposit date: | 2001-08-09 | | Release date: | 2001-09-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid.
Cell(Cambridge,Mass.), 85, 1996
|
|
1JNW
 
 | | Active Site Structure of E. coli pyridoxine 5'-phosphate Oxidase | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | di Salvo, M.L, Ko, T.P, Musayev, F.N, Raboni, S, Schirch, V, Safo, M.K. | | Deposit date: | 2001-07-25 | | Release date: | 2001-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Active site structure and stereospecificity of Escherichia coli pyridoxine-5'-phosphate oxidase.
J.Mol.Biol., 315, 2002
|
|
3Q36
 
 | |
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N6H
 
 | |
2NDM
 
 | | NMR solution structure of PawS Derived Peptide 21 (PDP-21) | | Descriptor: | PawS derived peptide 21 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
