5JST
 
 | | MBP fused MDV1 coiled coil | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein,Mitochondrial division protein 1, ... | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2016-05-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | ACCORD: an assessment tool to determine the orientation of homodimeric coiled-coils.
Sci Rep, 7, 2017
|
|
8TLX
 
 | |
8TLV
 
 | |
8TLW
 
 | |
5K94
 
 | | Deletion-Insertion Chimera of MBP with the Preprotein Cross-Linking Domain of the SecA ATPase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Maltose-binding periplasmic protein,Protein translocase subunit SecA,Maltose-binding periplasmic protein | | Authors: | Shilton, B.H, Hackett, J, Ghonaim, N. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of a polypeptide-binding site in the DEAD Motor of the SecA ATPase.
FEBS Lett., 591, 2017
|
|
4WVJ
 
 | | Crystal structure of the Type-I signal peptidase from Staphylococcus aureus (SpsB) in complex with an inhibitor peptide (pep3). | | Descriptor: | Maltose-binding periplasmic protein,Signal peptidase IB, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, inhibitor peptide (PEP3) | | Authors: | Young, P.G, Ting, Y.T, Baker, E.N. | | Deposit date: | 2014-11-05 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization.
IUCrJ, 3, 2016
|
|
4WMU
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 2 AT 1.55A | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Faiman, J.W. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WVH
 
 | | Crystal structure of the Type-I signal peptidase from Staphylococcus aureus (SpsB) in complex with a substrate peptide (pep1). | | Descriptor: | Maltose-binding periplasmic protein,Signal peptidase IB, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, substrate peptide (pep1) | | Authors: | Young, P.G, Ting, Y.T, Baker, E.N. | | Deposit date: | 2014-11-05 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization.
IUCrJ, 3, 2016
|
|
5JJ4
 
 | | Crystal Structure of a Variant Human Activation-induced Deoxycytidine Deaminase as an MBP fusion protein | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Single-stranded DNA cytosine deaminase, ZINC ION, ... | | Authors: | Pedersen, L.C, Goodman, M.F, Pham, P, Afif, S.A. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Structural analysis of the activation-induced deoxycytidine deaminase required in immunoglobulin diversification.
DNA Repair (Amst.), 43, 2016
|
|
8VG0
 
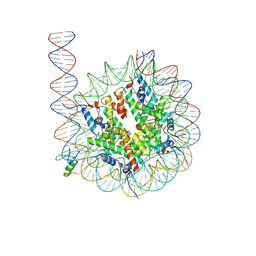 | | Cryo-EM structure of GATA4 in complex with ALBN1 nucleosome | | Descriptor: | DNA (159-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.R, Bai, Y. | | Deposit date: | 2023-12-22 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4.
Mol.Cell, 84, 2024
|
|
8VG1
 
 | |
5IMT
 
 | | Toxin receptor complex | | Descriptor: | CD59 glycoprotein, COPPER (II) ION, Intermedilysin, ... | | Authors: | Morton, C.J, Lawrence, S.L, Feil, S.C, Parker, M.W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7001 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
5II4
 
 | | Crystal structure of red abalone VERL repeat 1 with linker at 2.0 A resolution | | Descriptor: | Maltose-binding periplasmic protein,Vitelline envelope sperm lysin receptor, TRIETHYLENE GLYCOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5IQZ
 
 | | Crystal structure of N-terminal domain of Human SIRT7 | | Descriptor: | SIRT7 protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Thakur, K.G, Priyanka, A. | | Deposit date: | 2016-03-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Crystal structure of the N-terminal domain of human SIRT7 reveals a three-helical domain architecture
Proteins, 84, 2016
|
|
3J9P
 
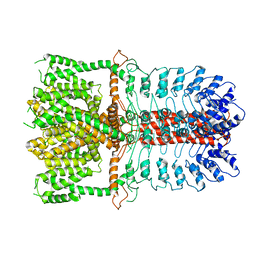 | | Structure of the TRPA1 ion channel determined by electron cryo-microscopy | | Descriptor: | Maltose-binding periplasmic protein, Transient receptor potential cation channel subfamily A member 1 chimera | | Authors: | Paulsen, C.E, Armache, J.-P, Gao, Y, Cheng, Y, Julius, D. | | Deposit date: | 2015-02-14 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structure of the TRPA1 ion channel suggests regulatory mechanisms.
Nature, 520, 2015
|
|
3JYR
 
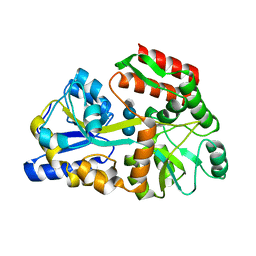 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Maltose-binding periplasmic protein | | Authors: | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | Deposit date: | 2009-09-22 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
8SVY
 
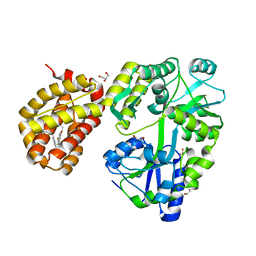 | | MBP-Mcl1 in complex with ligand 10 | | Descriptor: | (15P)-17-chloro-33-fluoro-12-[(2-methoxyethoxy)methyl]-5,14,22-trimethyl-28-oxa-9-thia-5,6,13,14,22-pentaazaheptacyclo[27.7.1.1~4,7~.0~11,15~.0~16,21~.0~20,24~.0~30,35~]octatriaconta-1(36),4(38),6,11(15),12,16,18,20,23,29(37),30,32,34-tridecaene-23-carboxylic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Macrocyclic Carbon-Linked Pyrazoles As Novel Inhibitors of MCL-1.
Acs Med.Chem.Lett., 14, 2023
|
|
4WMT
 
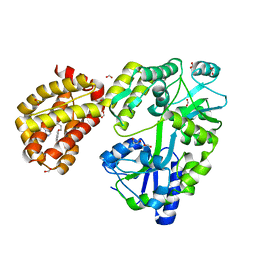 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 1 AT 2.35A | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-(1H-imidazol-1-yl)-4-methylpyridin-3-yl]-3-[3-(naphthalen-1-yloxy)propyl]-1-[2-oxo-2-(piperazin-1-yl)ethyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMV
 
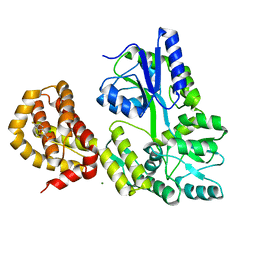 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 4 AT 2.4A | | Descriptor: | 3-chloro-6-fluoro-1-benzothiophene-2-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Moulin, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
5IIC
 
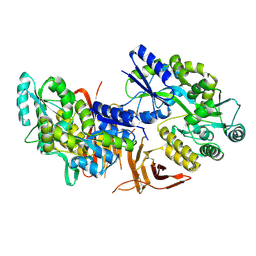 | | Crystal structure of red abalone VERL repeat 3 at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Vitelline envelope sperm lysin receptor, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
3LC8
 
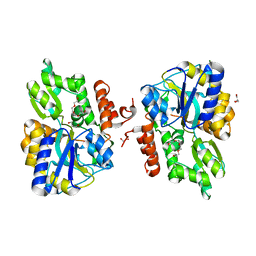 | |
3HPI
 
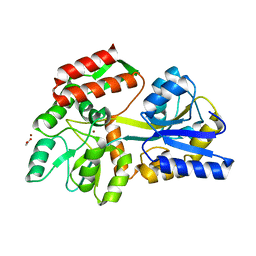 | |
8W8F
 
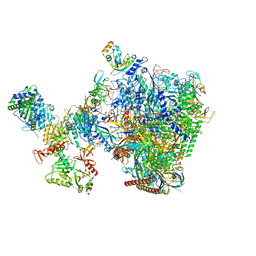 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
8VRS
 
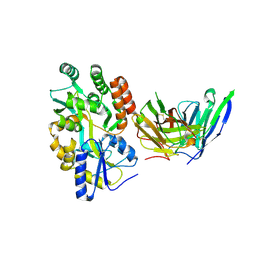 | | Mucin 16 peptide fused to MBP in complex with 4H11-scFv antibody | | Descriptor: | 4H11 scFv chain, Maltose/maltodextrin-binding periplasmic protein,Mucin-16 | | Authors: | Lee, K, Perry, K, Yeku, O.O, Spriggs, D.R. | | Deposit date: | 2024-01-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for antibody recognition of the proximal MUC16 ectodomain.
J Ovarian Res, 17, 2024
|
|
4XHS
 
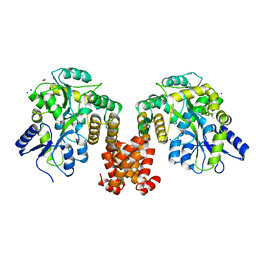 | | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 12, ... | | Authors: | Jin, T, Huang, M, Jiang, J, Xiao, T. | | Deposit date: | 2015-01-06 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction
To Be Published
|
|
