9DLV
 
 | | Cryo-EM structure of the human TREX-2.1 complex (LENG8/PCID2/DSS1) bound to DDX39B(UAP56) | | Descriptor: | 26S proteasome complex subunit SEM1, Leukocyte receptor cluster member 8, PCI domain-containing protein 2, ... | | Authors: | Clarke, B.P, Xie, Y, Ren, Y. | | Deposit date: | 2024-09-11 | | Release date: | 2025-09-17 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural mechanism of DDX39B regulation by human TREX-2 and a related complex in mRNP remodeling.
Nat Commun, 16, 2025
|
|
3E30
 
 | | Protein farnesyltransferase complexed with FPP and ethylene diamine inhibitor 4 | | Descriptor: | FARNESYL DIPHOSPHATE, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
7MEU
 
 | |
7M3Q
 
 | | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, ... | | Authors: | Chowdhury, A, Singer, A.U, Ogunjimi, A.A, Teyra, J, Zhang, W, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-03-18 | | Release date: | 2021-04-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV)
To be published
|
|
6NP0
 
 | | Cryo-EM structure of 5HT3A receptor in presence of granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular mechanism of setron-mediated inhibition of full-length 5-HT3Areceptor.
Nat Commun, 10, 2019
|
|
8Q3H
 
 | | Capra hircus reactive intermediate deaminase A mutant - A108D | | Descriptor: | 1,2-ETHANEDIOL, 2-iminobutanoate/2-iminopropanoate deaminase, GLYCEROL | | Authors: | Rizzi, G, Visentin, C, Di Pisa, F, Ricagno, S. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-26 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Site-directed mutagenesis reveals the interplay between stability, structure, and enzymatic activity in RidA from Capra hircus.
Protein Sci., 33, 2024
|
|
9D7N
 
 | | Human p38alpha MAP Kinase in complex with 1-Isobutyl-1H-indazole derivative; OSF267 | | Descriptor: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, 5-[(2,4-difluorophenyl)methyl]-N-[2-(dimethylamino)ethyl]-1-(2-methylpropyl)-1H-indazole-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Brunzelle, J.S, Shuvalova, L, Benetik, S.F, Roy, S.M, Watterson, D.M, Gobec, S. | | Deposit date: | 2024-08-16 | | Release date: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Targeting Neuroinflammation and Cognitive Decline: First-in-Class Dual Butyrylcholinesterase and p38 alpha Mitogen-Activated Protein Kinase Inhibitors.
J.Med.Chem., 68, 2025
|
|
4ZFV
 
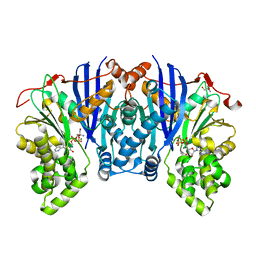 | | Lipomyces starkeyi levoglucosan kinase bound to ADP and magnesium. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Levoglucosan kinase, ... | | Authors: | Bacik, J.P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Producing Glucose 6-Phosphate from Cellulosic Biomass: STRUCTURAL INSIGHTS INTO LEVOGLUCOSAN BIOCONVERSION.
J.Biol.Chem., 290, 2015
|
|
8QH0
 
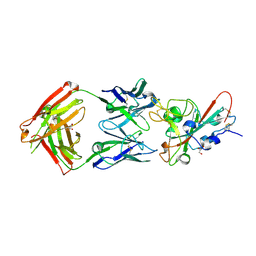 | | Crystal structure of the SARS-CoV-2 RBD with the antibody Cv2.3194 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cv2.3194 Heavy chain, GLYCEROL, ... | | Authors: | Fernandez, I, Rey, F.A. | | Deposit date: | 2023-09-06 | | Release date: | 2024-06-19 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Broad sarbecovirus neutralization by combined memory B cell antibodies to ancestral SARS-CoV-2.
Iscience, 27, 2024
|
|
4PC4
 
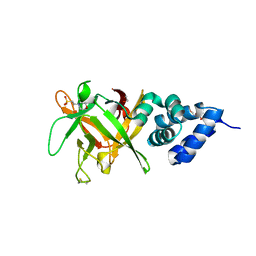 | | Bombyx mori lipoprotein 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 30K lipoprotein, ... | | Authors: | Pietrzyk, A.J, Bujacz, A, Jaskolski, M, Bujacz, G. | | Deposit date: | 2014-04-14 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bombyx mori lipoprotein 6: comparative structural analysis of the 30-kDa lipoprotein family.
Plos One, 9, 2014
|
|
5GVE
 
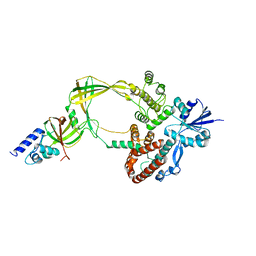 | | Human TOP3B-TDRD3 complex | | Descriptor: | DNA topoisomerase 3-beta-1, MAGNESIUM ION, Tudor domain-containing protein 3 | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.606 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
2XL3
 
 | | WDR5 IN COMPLEX WITH AN RBBP5 PEPTIDE AND HISTONE H3 PEPTIDE | | Descriptor: | GLYCEROL, HISTONE H3.1, RETINOBLASTOMA-BINDING PROTEIN 5, ... | | Authors: | Odho, Z, Southall, S.M, Wilson, J.R. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterisation of a Novel Wdr5 Binding Site that Recruits Rbbp5 Through a Conserved Motif and Enhances Methylation of H3K4 by Mll1.
J.Biol.Chem., 285, 2010
|
|
5FH0
 
 | | The structure of rat cytosolic PEPCK variant E89A complex with GTP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Johnson, T.A, Holyoak, T. | | Deposit date: | 2015-12-21 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Utilization of Substrate Intrinsic Binding Energy for Conformational Change and Catalytic Function in Phosphoenolpyruvate Carboxykinase.
Biochemistry, 55, 2016
|
|
4P7C
 
 | | Crystal structure of putative methyltransferase from Pseudomonas syringae pv. tomato | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative methyltransferase from Pseudomonas syringae pv. tomato
To Be Published
|
|
3G70
 
 | | Design and Preparation of Potent, Non-Peptidic, Bioavailable Renin Inhibitors | | Descriptor: | (1R,5S)-7-{4-[3-(2-chloro-3,6-difluorophenoxy)propyl]phenyl}-N-cyclopropyl-N-(2,3-dichlorobenzyl)-3,9-diazabicyclo[3.3.1]non-6-ene-6-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Bezencon, O, Bur, D, Prade, L, Weller, T, Boss, C, Fischli, W. | | Deposit date: | 2009-02-09 | | Release date: | 2009-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Preparation of Potent, Nonpeptidic, Bioavailable Renin Inhibitors
J.Med.Chem., 52, 2009
|
|
9DS0
 
 | | Crystal structure of sphA with MeVGQ | | Descriptor: | (2E,3E)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pent-3-enoic acid, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Xi, W, Hai, Y. | | Deposit date: | 2024-09-26 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | strucutre of PLP-dependent enzyme sphA with MeVGQ
To Be Published
|
|
3EO9
 
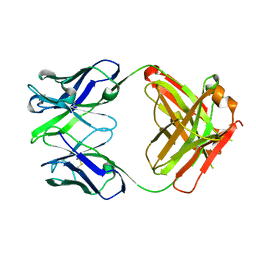 | | Crystal structure the Fab fragment of Efalizumab | | Descriptor: | Efalizumab Fab fragment, heavy chain, light chain | | Authors: | Li, S, Ding, J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efalizumab binding to the LFA-1 alphaL I domain blocks ICAM-1 binding via steric hindrance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7MJJ
 
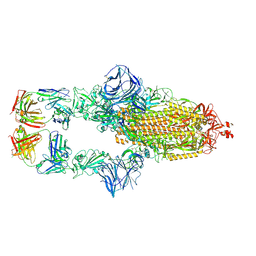 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
1L62
 
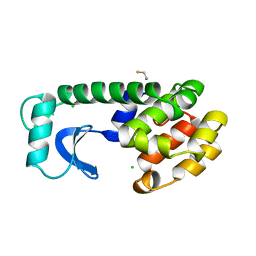 | |
3V4N
 
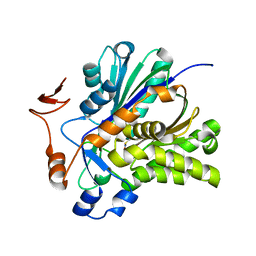 | | The Biochemical and Structural Basis for Inhibition of Enterococcus faecalis HMG-CoA Synthatse, mvaS, by Hymeglusin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HMG-CoA synthase | | Authors: | Skaff, D.A, Ramyar, K.X, McWhorter, W.J, Geisbrecht, B.V, Miziorko, H.M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and structural basis for inhibition of Enterococcus faecalis hydroxymethylglutaryl-CoA synthase, mvaS, by hymeglusin.
Biochemistry, 51, 2012
|
|
9D5P
 
 | |
6ASE
 
 | | KRAS mutant-A59G in GDP-bound | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Westover, K, Lu, J. | | Deposit date: | 2017-08-24 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | KRAS Switch Mutants D33E and A59G Crystallize in the State 1 Conformation.
Biochemistry, 57, 2018
|
|
5CZB
 
 | | HCV NS5B IN COMPLEX WITH LIGAND IDX17119-5 | | Descriptor: | 1-[4-(7-amino-5-methylpyrazolo[1,5-a]pyrimidin-2-yl)phenyl]-3-{[(R)-(2,4-dimethylphenyl)(methoxy)phosphoryl]amino}-1H-pyrazole-4-carboxylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pierra, C, Dousson, C, Augustin, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Synthesis of potent and broad genotypically active NS5B HCV non-nucleoside inhibitors binding to the thumb domain allosteric site 2 of the viral polymerase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
9D5H
 
 | |
9D5E
 
 | |
