6IRY
 
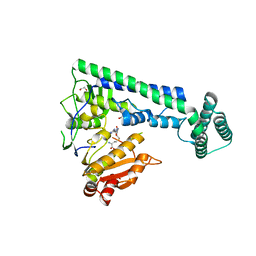 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
7ZYK
 
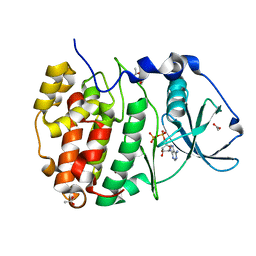 | | Compound 9 Bound to CK2alpha | | Descriptor: | 2-(5-bromanyl-6-chloranyl-1~{H}-indol-3-yl)ethanenitrile, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
7ZUR
 
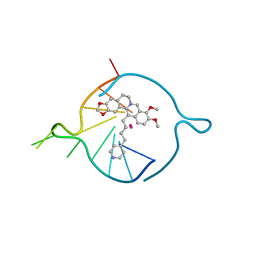 | | Four carbons pendant pyridine derivative of the natural alkaloid Berberine as Human Telomeric G-quadruplex Binder | | Descriptor: | 16,17-dimethoxy-21-(4-pyridin-4-ylbutyl)-5,7-dioxa-13$l^{4}-azapentacyclo[11.8.0.0^{2,10}.0^{4,8}.0^{15,20}]henicosa-1(21),2(10),3,8,13,15,17,19-octaene, DNA TAGGGTTAGGGT, POTASSIUM ION | | Authors: | Bazzicalupi, C, Ferraroni, M, Gratteri, P, Papi, F. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Efficiency of 13-Pyridylalkyl Berberine Derivatives to Human Telomeric G-Quadruplexes Binding: Spectroscopic, Solid State and In Silico Analysis.
Int J Mol Sci, 23, 2022
|
|
4Y5B
 
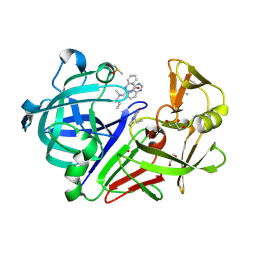 | | Endothiapepsin in complex with fragment 261 | | Descriptor: | 2-({[(1S)-1-cyclopropylethyl](methyl)amino}methyl)quinazolin-4(3H)-one, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
to be published
|
|
6V51
 
 | | Spin-labeled T4 Lysozyme (9/131FnbY)-(4-Amino-TEMPO) | | Descriptor: | 4-amino-2,2,6,6-tetramethylpiperidin-1-ol, Endolysin | | Authors: | Liu, J, Morizumi, T, Ou, W.L, Wang, L, Ernst, O.P. | | Deposit date: | 2019-12-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents.
J.Am.Chem.Soc., 142, 2020
|
|
5KD5
 
 | |
5HZL
 
 | |
4MSG
 
 | | Crystal structure of tankyrase 1 with compound 22 | | Descriptor: | 3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]-N-[trans-4-(5-phenyl-1,3,4-oxadiazol-2-yl)cyclohexyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
3OVA
 
 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, CCA-adding enzyme, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
5HZG
 
 | | The crystal structure of the strigolactone-induced AtD14-D3-ASK1 complex | | Descriptor: | (2Z)-2-methylbut-2-ene-1,4-diol, F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A, ... | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
5BXL
 
 | | Yeast 20S proteasome beta2-G170A mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defective immuno- and thymoproteasome assembly causes severe immunodeficiency.
Sci Rep, 8, 2018
|
|
3ON2
 
 | | Structure of a protein with unknown function from Rhodococcus sp. RHA1 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Probable transcriptional regulator, SULFATE ION | | Authors: | Fan, Y, Evdokimova, E, Egorova, O, Savchenko, A, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of a protein with unknown function from Rhodococcus sp. RHA1
To be Published
|
|
5HAP
 
 | | OXA-48 beta-lactamase - S70A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Stojanoski, V, Adamski, C.J, Hu, L, Mehta, S.C, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Removal of the Side Chain at the Active-Site Serine by a Glycine Substitution Increases the Stability of a Wide Range of Serine beta-Lactamases by Relieving Steric Strain.
Biochemistry, 55, 2016
|
|
5KHB
 
 | | Structure of Phenol-soluble modulin Alpha1 | | Descriptor: | PSM Alpha1 | | Authors: | Towle, K.M, Lohans, C.T, Acedo, J.Z, Miskolzie, M, van Belkum, M.J, Vederas, J.C. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Phenol-Soluble Modulins alpha 1, alpha 3, and beta 2, Virulence Factors from Staphylococcus aureus.
Biochemistry, 55, 2016
|
|
2BVD
 
 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | (3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)piperidin-3-yl beta-D-glucopyranoside, ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-27 | | Release date: | 2005-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
6UZK
 
 | |
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2024-07-24 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
5FGJ
 
 | | Structure of tetrameric rat phenylalanine hydroxylase, residues 1-453 | | Descriptor: | FE (III) ION, MAGNESIUM ION, Phenylalanine-4-hydroxylase | | Authors: | Taylor, A.B, Roberts, K.M, Fitzpatrick, P.F. | | Deposit date: | 2015-12-20 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Domain Movements upon Activation of Phenylalanine Hydroxylase Characterized by Crystallography and Chromatography-Coupled Small-Angle X-ray Scattering.
J.Am.Chem.Soc., 138, 2016
|
|
6J27
 
 | | Crystal structure of the branched-chain polyamine synthase from Thermus thermophilus (Tth-BpsA) in complex with N4-aminopropylspermidine and 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
7GG9
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ANT-DIA-62e4526e-1 (Mpro-x12204) | | Descriptor: | 2-(1H-1,2,3-benzotriazol-1-yl)-1-(4-methylpiperidin-1-yl)ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6UW2
 
 | | Clotrimazole bound complex of Acanthamoeba castellanii CYP51 | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, GUANIDINE, Obtusifoliol 14alphademethylase, ... | | Authors: | Sharma, V, Podust, L.M. | | Deposit date: | 2019-11-04 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Domain-Swap Dimerization of Acanthamoeba castellanii CYP51 and a Unique Mechanism of Inactivation by Isavuconazole.
Mol.Pharmacol., 98, 2020
|
|
6UWV
 
 | | BACE-1 in complex with compound #34 | | Descriptor: | (4aR,7aR)-7a-[(1R,2R)-2-(2-{[(1R,2R)-2-methylcyclopropyl]methoxy}propan-2-yl)cyclopropyl]-6-(pyrimidin-2-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Stout, S.L. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6V1K
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to BI7273 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-[(dimethylamino)methyl]-3,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6CBN
 
 | | x-ray structure of NeoB from streptomyces fradiae in complex with PLP and neomycin (as the external aldimine) at pH 7.5 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-[(3-O-{2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranosyl}-beta-D-ribofuranosyl)oxy]-3-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, Neamine transaminase NeoN | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
7XVI
 
 | |
