6V5N
 
 | | EGFR(T790M/V948R) in complex with LN2084 | | Descriptor: | 3-[4-(4-fluorophenyl)-5-(2-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)-1H-imidazol-2-yl]propan-1-ol, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
3JSF
 
 | |
6V8D
 
 | | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators | | Descriptor: | (3Z)-3-iminocyclohex-1-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Zhu, W, Doubleday, P.T, Catlin, D.S, Weerawarna, P, Butrin, A, Shen, S, Kelleher, N.L, Liu, D, Silverman, R.B. | | Deposit date: | 2019-12-10 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators
To Be Published
|
|
3E9B
 
 | |
5WGI
 
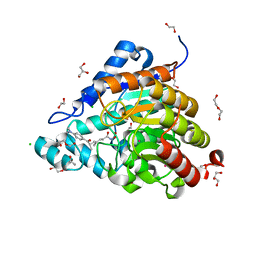 | |
9C55
 
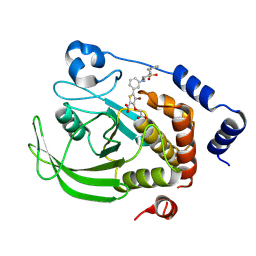 | | Crystal structure of human PTPN2 in complex with active site inhibitor | | Descriptor: | 5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Bester, S.M, Linwood, R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Enhancing the apo protein tyrosine phosphatase non-receptor type 2 crystal soaking strategy through inhibitor-accessible binding sites.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9HCC
 
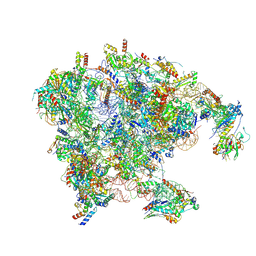 | | Mouse mitoribosome large subunit assembly intermediate (without uL16m) bound to MRM3-dimer, DDX28 and the MALSU-L0R8F8-mt-ACP complex, State A1 (SAMC knock-out) | | Descriptor: | 16S rRNA (1584-MER), Acyl carrier protein, mitochondrial, ... | | Authors: | Singh, V, Rorbach, J, Freyer, C, Amunts, A, Wredenberg, A. | | Deposit date: | 2024-11-08 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (5.67 Å) | | Cite: | The mitochondrial methylation potential gates mitoribosome assembly.
Nat Commun, 16, 2025
|
|
9C8U
 
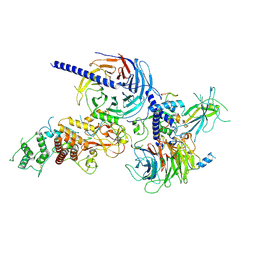 | | Human PRC2 - RvLEAM (short) (1:6 molar ratio), cross-linked 10 min | | Descriptor: | Isoform 2 of Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, Polycomb protein SUZ12, ... | | Authors: | Abe, K.M, Li, G, He, Q, Grant, T, Lim, C. | | Deposit date: | 2024-06-13 | | Release date: | 2024-09-04 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Small LEA proteins mitigate air-water interface damage to fragile cryo-EM samples during plunge freezing.
Nat Commun, 15, 2024
|
|
7HHR
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z57744712 | | Descriptor: | 1,2-ETHANEDIOL, Hepatoma-derived growth factor-related protein 2, N,N-dimethylmorpholine-4-sulfonamide, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
9CBK
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 6 in Complex with p-Aminomethyl Phenylthioketone | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(aminomethyl)phenyl]-2-sulfanylethan-1-one, Hdac6 protein, ... | | Authors: | Goulart Stollmaier, J, Watson, P.R, Christianson, D.W. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Design, Synthesis, and Structural Evaluation of Acetylated Phenylthioketone Inhibitors of HDAC10.
Acs Med.Chem.Lett., 15, 2024
|
|
4H07
 
 | | Complex of G65T Myoglobin with Phenol in its Proximal Cavity | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PHENOL, ... | | Authors: | Lebioda, L, Lovelace, L.L, Celeste, L.R, Huang, X, Wang, C, Shengfang, S, Dawson, J.H. | | Deposit date: | 2012-09-07 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Complex of myoglobin with phenol bound in a proximal cavity.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
9CF0
 
 | | Parasitella parasitica Fanzor (PpFz) State 1 | | Descriptor: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
7HN5
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z45617795 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N-(2-phenylethyl)methanesulfonamide, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HNB
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z31478538 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N-Benzyl-2-methoxyacetamide, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5KPP
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[(3R)-3-methyl-4-[2,2,2-tris(fluoranyl)ethyl]piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
7HNZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z1245793018 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N,N-dimethylpyridin-4-amine, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HO2
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z730649594 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N-[2-(4-hydroxyphenyl)ethyl]pyridine-2-carboxamide, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HLR
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z1430613393 | | Descriptor: | 1,2-ETHANEDIOL, 3-fluoro-N-(3-hydroxy-4-methylphenyl)benzamide, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
8JLK
 
 | | Ulotaront(SEP-363856)-bound mTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
7HM0
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z374427992 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N-{[2-(dimethylamino)pyridin-4-yl]methyl}acetamide, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
8JLO
 
 | | Ulotaront(SEP-363856)-bound hTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6G7F
 
 | | Yeast 20S proteasome in complex with Cystargolide B | | Descriptor: | CHLORIDE ION, Cystargolide B- bound form, MAGNESIUM ION, ... | | Authors: | Groll, M, Tello-Aburto, R. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-12 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, synthesis, and evaluation of cystargolide-based beta-lactones as potent proteasome inhibitors.
Eur J Med Chem, 157, 2018
|
|
9HCE
 
 | | Mouse mitoribosome large subunit assembly intermediate (without uL16m) bound to MRM3 dimer and the MALSU-L0R8F8-mt-ACP complex, State A3 (SAMC knock out) | | Descriptor: | 16S rRNA (1584-MER), Acyl carrier protein, mitochondrial, ... | | Authors: | Singh, V, Rorbach, J, Freyer, C, Amunts, A, Wredenberg, A. | | Deposit date: | 2024-11-08 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (7.31 Å) | | Cite: | The mitochondrial methylation potential gates mitoribosome assembly.
Nat Commun, 16, 2025
|
|
9L5R
 
 | | Cryo-EM structure of the thermophile spliceosome (state ILS) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Cell cycle control protein, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
7BZL
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-04-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
