5D19
 
 | |
4L2H
 
 | | Structure of a catalytically inactive PARG in complex with a poly-ADP-ribose fragment | | Descriptor: | Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Brassington, A, Dunstan, M.S, Leys, D. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities.
Nat Commun, 4, 2013
|
|
6JX6
 
 | | Tetrameric form of Smac | | Descriptor: | Diablo homolog, mitochondrial | | Authors: | Sivaraman, J, Singh, S, Ng, J, Nayak, D. | | Deposit date: | 2019-04-22 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activitiesin vitro.
J.Biol.Chem., 294, 2019
|
|
2YFW
 
 | |
1J2W
 
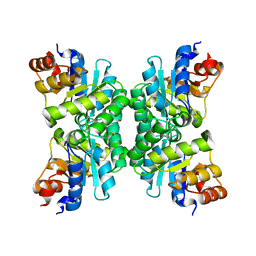 | | Tetrameric Structure of aldolase from Thermus thermophilus HB8 | | Descriptor: | Aldolase protein | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3JAO
 
 | | Ciliary microtubule doublet | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Maheshwari, A, Obbineni, J.M, Bui, K.H, Shibata, K, Toyoshima, Y.Y, Ishikawa, T. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | alpha- and beta-Tubulin Lattice of the Axonemal Microtubule Doublet and Binding Proteins Revealed by Single Particle Cryo-Electron Microscopy and Tomography.
Structure, 23, 2015
|
|
8TJU
 
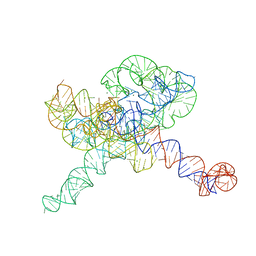 | | Tetrahymena Ribozyme scaffolded TABV xrRNA | | Descriptor: | DNA/RNA (416-MER), MAGNESIUM ION | | Authors: | Langeberg, C.J, Kieft, J.S. | | Deposit date: | 2023-07-24 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | A generalizable scaffold-based approach for structure determination of RNAs by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7PQ8
 
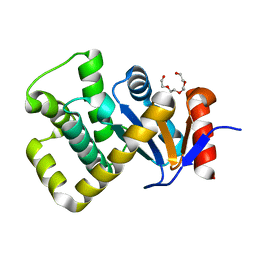 | | Crystal structure of Campylobacter jejuni DsbA1 | | Descriptor: | TETRAETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Orlikowska, M, Bocian-Ostrzycka, K.M, Banas, A.M, Jagusztyn-Krynicka, E.K. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
7PQ7
 
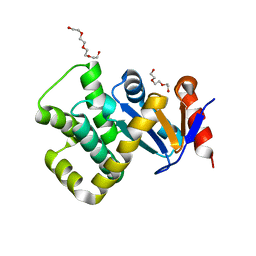 | | Crystal structure of Campylobacter jejuni DsbA1 | | Descriptor: | TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Wilk, P, Orlikowska, M, Banas, A.M, Bocian-Ostrzycka, K.M, Jagusztyn-Krynicka, E.K. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
5OJX
 
 | | Crystal structure of regulator protein 2 (PamR2) from the pamamycin biosynthetic gene cluster of Streptomyces alboniger | | Descriptor: | TetR family transcription regulator | | Authors: | Schmelz, S, Rebets, Y, Luzhetskyy, A, Scrima, A. | | Deposit date: | 2017-07-24 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, development and application of whole-cell based antibiotic-specific biosensor.
Metab. Eng., 47, 2018
|
|
8X5N
 
 | | Tetramer Gabija with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, Gabija protein GajB, ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
6PNK
 
 | |
6N4Z
 
 | | Tetragonal thermolysin (with 50% xylose) plunge cooled in liquid nitrogen to 77 K | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION, ... | | Authors: | Juers, D.H, Harrison, K, Wu, B. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A comparison of gas stream cooling and plunge cooling of macromolecular crystals.
J.Appl.Crystallogr., 52, 2019
|
|
6N4W
 
 | | Tetragonal thermolysin (with 50% xylose) cryocooled in a nitrogen gas stream to 100 K | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION, ... | | Authors: | Juers, D.H, Harrison, K, Wu, B. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A comparison of gas stream cooling and plunge cooling of macromolecular crystals.
J.Appl.Crystallogr., 52, 2019
|
|
8DOT
 
 | |
7B8I
 
 | | Tetragonal structure of human protein kinase CK2 catalytic subunit in complex with a heparin oligo saccharide | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Schnitzler, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the design of bisubstrate inhibitors of protein kinase CK2 provided by complex structures with the substrate-competitive inhibitor heparin.
Eur.J.Med.Chem., 214, 2021
|
|
6VE3
 
 | | Tetradecameric PilQ from Pseudomonas aeruginosa | | Descriptor: | Fimbrial assembly protein PilQ | | Authors: | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
4XF2
 
 | | Tetragonal structure of Arp2/3 complex | | Descriptor: | Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, Actin-related protein 2/3 complex subunit 2, ... | | Authors: | Jurgenson, C.T, Pollard, T.P. | | Deposit date: | 2014-12-25 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Crystals of the Arp2/3 complex in two new space groups with structural information about actin-related protein 2 and potential WASP binding sites.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4R4G
 
 | |
6FZZ
 
 | | Crystal structure of BSE31 (BSPA14S_RS05060 gene product) from Lyme disease agent Borrelia (Borreliella) spielmanii | | Descriptor: | TETRAETHYLENE GLYCOL, Virulent strain associated lipoprotein | | Authors: | Brangulis, K, Akopjana, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-03-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BBE31 from the Lyme disease agent Borrelia burgdorferi, known to play an important role in successful colonization of the mammalian host, shows the ability to bind glutathione.
Biochim Biophys Acta Gen Subj, 1864, 2019
|
|
3D1B
 
 | | Tetragonal crystal structure of Tas3 C-terminal alpha motif | | Descriptor: | RNA-induced transcriptional silencing complex protein tas3 | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.
Mol.Cell, 34, 2009
|
|
7XZO
 
 | | Formate-tetrahydrofolate ligase in complex with ATP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of FtfL as a novel target of berberine in intestinal bacteria.
Bmc Biol., 21, 2023
|
|
7XZN
 
 | |
7XZP
 
 | |
7Z14
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with a short-chain neurotoxin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Nys, M.A.E.M, Zarkadas, E, Ulens, C, Nury, H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The molecular mechanism of snake short-chain alpha-neurotoxin binding to muscle-type nicotinic acetylcholine receptors.
Nat Commun, 13, 2022
|
|
