6YOO
 
 | | Structure of SAMM50 LIR bound to GABARAPL1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Wenxin, Z, Johansen, T, Tooze, S, Abudu, Y.P, Lamark, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structure of SAMM50 LIR bound to GABARAPL1
To Be Published
|
|
7BOO
 
 | | Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form | | Descriptor: | Alpha-1,2-mannosyltransferase (Ktr4), putative, GLYCEROL, ... | | Authors: | Hira, D, Onoue, T, Oka, T. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the core-mannan biosynthesis of cell wall fungal-type galactomannan in Aspergillus fumigatus .
J.Biol.Chem., 295, 2020
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
8RVB
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 8 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[2-(1~{H}-1,2,3-triazol-4-yl)ethylsulfanylmethyl]oxolane-3,4-diol, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
7BOP
 
 | | Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form | | Descriptor: | Alpha-1,2-mannosyltransferase (Ktr4), putative, GLYCEROL, ... | | Authors: | Hira, D, Onoue, T, Oka, T. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the core-mannan biosynthesis of cell wall fungal-type galactomannan in Aspergillus fumigatus .
J.Biol.Chem., 295, 2020
|
|
5JV1
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor CL-08-066 | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION, [(1R)-1-{[6-(3-chloro-4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}-2-(3-fluorophenyl)ethyl]phosphonic acid | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
6TTE
 
 | | Beta-galactosidase in complex with PETG | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
6THM
 
 | | Linalool Dehydratase Isomerase M125A mutant | | Descriptor: | 1,2-ETHANEDIOL, Linalool dehydratase-isomerase protein LDI, MALONATE ION | | Authors: | Cuetos, A, Zukic, E, Danesh-Azari, H.R, Grogan, G. | | Deposit date: | 2019-11-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Mutational Analysis of Linalool Dehydratase Isomerase Suggests That Alcohol and Alkene Transformations Are Catalyzed Using Noncovalent Mechanisms
Acs Catalysis, 2020
|
|
6QE9
 
 | | The X-ray structure of the adduct formed in the reaction between bovine pancreatic ribonuclease and complex I, a pentacoordinate Pt(II) compound containing 2,9-dimethyl-1,10-phenanthroline, dimethylfumarate, methyl and iodine as ligands | | Descriptor: | Ribonuclease pancreatic, SULFATE ION, pentacoordinate Pt(II) compound | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2019-01-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Reaction with Proteins of a Five-Coordinate Platinum(II) Compound.
Int J Mol Sci, 20, 2019
|
|
6CC7
 
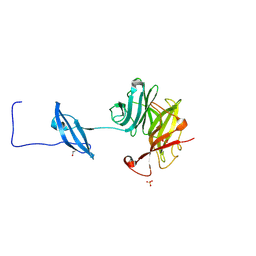 | | Structure of Fungal GH62 from Thielavia terretris | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycoside hydrolase family 62 protein, ... | | Authors: | Camargo, S, Mulinari, E.J, Almeida, L.R, Muniz, J.R.C. | | Deposit date: | 2018-02-06 | | Release date: | 2019-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Fungal GH62 from Thielavia terretris
To Be Published
|
|
3EC7
 
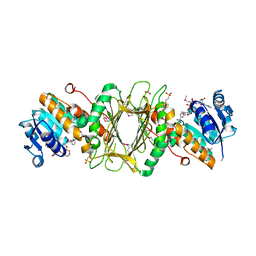 | | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETIC ACID, ... | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2
To be Published
|
|
3UF1
 
 | | Crystal Structure of Vimentin (fragment 144-251) from Homo sapiens, Northeast Structural Genomics Consortium Target HR4796B | | Descriptor: | Vimentin | | Authors: | Kuzin, A, Abashidze, M, Vorobiev, S.M, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography.
J.Biol.Chem., 287, 2012
|
|
2XH6
 
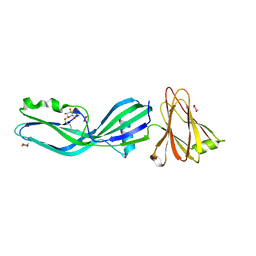 | | Clostridium perfringens enterotoxin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, HEAT-LABILE ENTEROTOXIN B CHAIN, octyl beta-D-glucopyranoside | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, MCClane, B.A, Basak, A.K. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
4Z2S
 
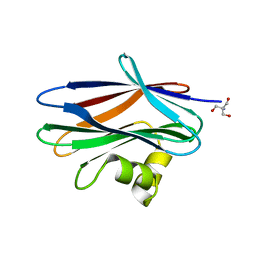 | | The crystal structure of Sclerotium Rolfsii lectin variant 2 (SSR2) in complex with N-acetyl-glucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
5W8Y
 
 | |
4Z4A
 
 | |
8B0O
 
 | | Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3 | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{R})-3-[(2~{R})-3-[[(2~{R})-1-[[(2~{R})-1-[[(2~{R})-6-[(2-aminophenyl)carbonylamino]-1-azanyl-1-oxidanylidene-hexan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8HYB
 
 | |
8D5N
 
 | | Crystal structure of Ld-HF10 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2022-06-05 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide Centric V beta Specific Germline Contacts Shape a Specialist T Cell Response.
Front Immunol, 13, 2022
|
|
8X2J
 
 | |
5M36
 
 | | The molecular tweezer CLR01 stabilizes a disordered protein-protein interface | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein zeta/delta, BENZOIC ACID, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2016-10-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Molecular Tweezer CLR01 Stabilizes a Disordered Protein-Protein Interface.
J. Am. Chem. Soc., 139, 2017
|
|
7BT5
 
 | | Crystal structure of plasmodium LysRS complexing with an antitumor compound | | Descriptor: | LYSINE, Lysine--tRNA ligase, N4-[2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-N2-(2-propan-2-ylsulfonylphenyl)-1,3,5-triazine-2,4-diamine | | Authors: | Zhou, J, Wang, J, Fang, P. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Inhibition of Plasmodium falciparum Lysyl-tRNA synthetase via an anaplastic lymphoma kinase inhibitor.
Nucleic Acids Res., 48, 2020
|
|
5U2P
 
 | | The crystal structure of Tp0737 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Norgard, M.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Functional clues from the crystal structure of an orphan periplasmic ligand-binding protein from Treponema pallidum.
Protein Sci., 26, 2017
|
|
8XLO
 
 | |
5OO8
 
 | | Streptomyces PAC13 (H42Q) with uridine | | Descriptor: | 1,2-ETHANEDIOL, Putative cupin_2 domain-containing isomerase, URIDINE | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
