1ZBP
 
 | | X-Ray Crystal Structure of Protein VPA1032 from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR44 | | Descriptor: | hypothetical protein VPA1032 | | Authors: | Forouhar, F, Yong, W, Vorobiev, S.M, Ciao, M, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3SRN
 
 | | STRUCTURAL CHANGES THAT ACCOMPANY THE REDUCED CATALYTIC EFFICIENCY OF TWO SEMISYNTHETIC RIBONUCLEASE ANALOGS | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | deMel, V.S.J, Martin, P.D, Doscher, M.S, Edwards, B.F.P. | | Deposit date: | 1991-05-20 | | Release date: | 1994-12-20 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural changes that accompany the reduced catalytic efficiency of two semisynthetic ribonuclease analogs.
J.Biol.Chem., 267, 1992
|
|
6GLA
 
 | | Crystal structure of JAK3 in complex with Compound 11 (FM481) | | Descriptor: | (~{E})-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2(6),4,7,11-pentaen-4-yl)furan-2-yl]prop-2-enenitrile, 1,2-ETHANEDIOL, 1-phenylurea, ... | | Authors: | Chaikuad, A, Forster, M, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Development, Optimization, and Structure-Activity Relationships of Covalent-Reversible JAK3 Inhibitors Based on a Tricyclic Imidazo[5,4- d]pyrrolo[2,3- b]pyridine Scaffold.
J. Med. Chem., 61, 2018
|
|
2P7T
 
 | | Crystal Structure of KcsA mutant | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, FAB-A, FAB-B, ... | | Authors: | Cordero-Morales, J.F, Vishwanath, J, Lewis, A, Valeria, V.R, Cortes, D.M, Roux, B, Perozo, E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-10-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of KcsA mutant
To be Published
|
|
7Y79
 
 | | Crystal structure of Cry78Aa | | Descriptor: | Toxin | | Authors: | Cao, B.B, Nie, Y.F, Wang, N.C, Guan, Z.Y, Zhang, D.L, Zhang, J. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of Cry78Aa from Bacillus thuringiensis provides insights into its insecticidal activity.
Commun Biol, 5, 2022
|
|
5KF9
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Holden, H.M, Tipton, P.A. | | Deposit date: | 2016-06-12 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
7K5X
 
 | |
4QB3
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with Olinone | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-[4-(1-oxo-1,2,3,4-tetrahydro-5H-pyrido[4,3-b]indol-5-yl)butyl]acetamide | | Authors: | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | Deposit date: | 2014-05-06 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression.
Chem.Biol., 21, 2014
|
|
5IWM
 
 | | 2.5A structure of GSK945237 with S.aureus DNA gyrase and DNA. | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*TP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*AP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Miles, T.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3QRJ
 
 | | The crystal structure of human abl1 kinase domain T315I mutant in complex with DCC-2036 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
5GN5
 
 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17 | | Descriptor: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
4DKQ
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with DMJ-I-228 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(1S,2S)-2-carbamimidamido-2,3-dihydro-1H-inden-1-yl]-N'-(4-chloro-3-fluorophenyl)ethanediamide, ... | | Authors: | Kwon, Y.D, LaLonde, J.M, Jones, D.M, Sun, A.W, Courter, J.R, Soeta, T, Kobayashi, T, Princiotto, A.M, Wu, X, Mascola, J, Schon, A, Freire, E, Sodroski, J, Madani, N, Smith III, A.B, Kwong, P.D. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors.
J.Med.Chem., 55, 2012
|
|
7K63
 
 | |
7K61
 
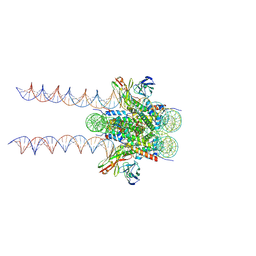 | | Cryo-EM structure of 197bp nucleosome aided by scFv | | Descriptor: | DNA (197-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.-R, Bai, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms.
Mol.Cell, 81, 2021
|
|
6QX2
 
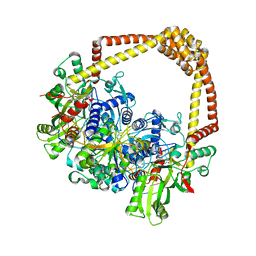 | | 3.4A structure of benzoisoxazole 3 with S.aureus DNA gyrase and DNA | | Descriptor: | (2~{R})-2-[[5-(2-chlorophenyl)-1,2-benzoxazol-3-yl]oxy]-2-phenyl-ethanamine, DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D. | | Deposit date: | 2019-03-06 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-guided design of antibacterials that allosterically inhibit DNA gyrase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3TS6
 
 | |
6IZ3
 
 | |
6CNE
 
 | | Selenomethionine variant (V29SeM) of protein GB1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, PHOSPHATE ION | | Authors: | Chen, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
5UNM
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, substrate free form with flexible loop | | Descriptor: | ATP-utilizing enzyme of the PP-loopsuperfamily, PHOSPHATE ION | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-01-31 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7YG3
 
 | | Crystal structure of HLA-B*13:01 | | Descriptor: | ARG-GLN-ASP-ILE-LEU-ASP-LEU-TRP-ILE, Beta-2-microglobulin, MHC class I antigen | | Authors: | Wang, H.S, Ouyang, S.Y. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional and structural characteristics of HLA-B*13:01-mediated specific T cells reaction in dapsone-induced drug hypersensitivity.
J.Biomed.Sci., 29, 2022
|
|
5KGA
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant D287N, in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
6J3G
 
 | | Crystal structure of an apo form of the glutathione S-transferase, CsGST83044, of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glutathione S-transferase, ... | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of key residues for activities of atypical glutathione S-transferase of Ceriporiopsis subvermispora, a selective degrader of lignin in woody biomass, by crystallography and functional mutagenesis.
Int.J.Biol.Macromol., 132, 2019
|
|
3PC0
 
 | | Crystal structure of the mutant V155S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
7K5Y
 
 | |
2POG
 
 | | Benzopyrans as Selective Estrogen Receptor b Agonists (SERBAs). Part 2: Structure Activity Relationship Studies on the Benzopyran Scaffold. | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-9-OL, Estrogen receptor | | Authors: | Richardson, T.I, Norman, B.H, Lugar, C.W, Jones, S.A, Wang, Y, Durbin, J.D, Krishnan, V, Dodge, J.A. | | Deposit date: | 2007-04-26 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 2: structure-activity relationship studies on the benzopyran scaffold.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
