1ANR
 
 | |
8W2F
 
 | | Plasmodium falciparum 20S proteasome bound to an inhibitor | | Descriptor: | (3S)-1-[(2-fluoroethoxy)acetyl]-N-{[(4P)-4-(6-methylpyridin-3-yl)-1,3-thiazol-2-yl]methyl}piperidine-3-carboxamide, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | Authors: | Han, Y, Deng, X, Ray, S, Chen, Z, Phillips, M. | | Deposit date: | 2024-02-20 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Identification of potent and reversible piperidine carboxamides that are species-selective orally active proteasome inhibitors to treat malaria.
Cell Chem Biol, 31, 2024
|
|
4ZGY
 
 | |
8Z07
 
 | |
4RHI
 
 | |
8DGM
 
 | | 14-3-3 epsilon bound to phosphorylated PEAK1 (pT1165) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Inactive tyrosine-protein kinase PEAK1 | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
6E85
 
 | | 1.25 Angstrom Resolution Crystal Structure of 4-hydroxythreonine-4-phosphate Dehydrogenase from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, D-threonate 4-phosphate dehydrogenase, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
8ANK
 
 | |
5KFM
 
 | |
7UUO
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA H135A mutant, complex with tobramycin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, COENZYME A, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7FZ1
 
 | | Crystal Structure of human FABP4 in complex with 3-[(E)-anilino-(2-oxo-1H-indol-3-ylidene)methyl]sulfanylpropanoic acid | | Descriptor: | 3-{[(E)-anilino(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]sulfanyl}propanoic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Mischke, S, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5GUL
 
 | | Crystal structure of Tris/PPix2/Mg2+ bound form of cyclolavandulyl diphosphate synthase (CLDS) from Streptomyces sp. CL190 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cyclolavandulyl diphosphate synthase, MAGNESIUM ION, ... | | Authors: | Tomita, T, Kobayashi, M, Nishiyama, M, Kuzuyama, T. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure and Mechanism of the Monoterpene Cyclolavandulyl Diphosphate Synthase that Catalyzes Consecutive Condensation and Cyclization.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5KGA
 
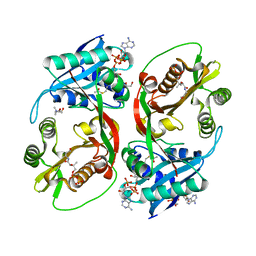 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant D287N, in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
8IWC
 
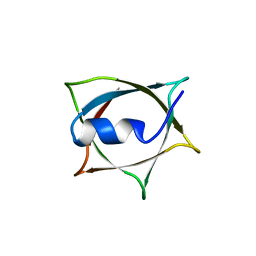 | | Crystal structure of Q9PR55 at pH 6.0 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8DGP
 
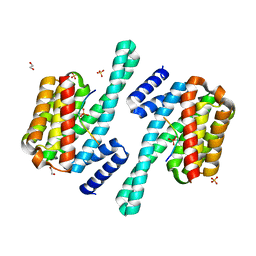 | | 14-3-3 epsilon bound to phosphorylated PEAK3 (pS69) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Phosphorylated PEAK3 (pS69) peptide, ... | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
8IWA
 
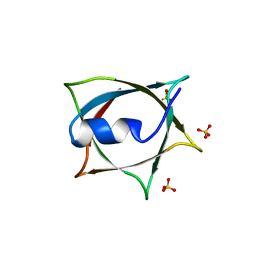 | | Crystal structure of Q9PR55 at pH 6.5 | | Descriptor: | SULFATE ION, Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8RQK
 
 | |
9DM5
 
 | | Product-Bound mannosyltransferase PimE | | Descriptor: | (2S)-1-{[(S)-{[(1S,2R,3R,4S,5S,6R)-2-[(6-O-hexadecanoyl-beta-L-gulopyranosyl)oxy]-3,4,5-trihydroxy-6-{[beta-D-mannopyranosyl-(1->2)-alpha-D-mannopyranosyl-(1->6)-beta-D-mannopyranosyl-(1->6)-alpha-D-mannopyranosyl]oxy}cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}-3-(hexadecanoyloxy)propan-2-yl 10-methyloctadecanoate, MONO-TRANS, OCTA-CIS DECAPRENYL-PHOSPHATE, ... | | Authors: | Liu, Y. | | Deposit date: | 2024-09-12 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
8IWB
 
 | | Crystal structure of Q9PR55 at pH 7.5 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
7XYJ
 
 | | Structure of WSSV thymidylate synthase in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Ma, Q, Liu, C, Zang, K. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Structure of WSSV thymidylate synthase in complex with dUMP
To Be Published
|
|
8D9Y
 
 | | Crystal structure of Taipan alpha-neurotoxin in complex with Centi-LNX-D09 antibody | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Centi-LNX-D09 Fab heavy chain, ... | | Authors: | Pletnev, S, Verardi, R, Tully, E.S, Glanville, J, Kwong, P.D. | | Deposit date: | 2022-06-12 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Venom protection by antibody from a snakebite hyperimmune subject
To Be Published
|
|
6Z4B
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with Osimertinib and EAI045 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, SULFATE ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complex Crystal Structures of EGFR with Third-Generation Kinase Inhibitors and Simultaneously Bound Allosteric Ligands.
Acs Med.Chem.Lett., 11, 2020
|
|
9D7J
 
 | | Clostridium acetobutylicum alcohol dehydrogenase bound to NADP+ | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Madzelan, P, Wilson, M.A. | | Deposit date: | 2024-08-16 | | Release date: | 2025-07-02 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Is a Malleable Active Site Loop the Key to High Substrate Promiscuity? Hybrid, Biocatalytic Route to Structurally Diverse Taxoid Side Chains with Remarkable Dual Stereocontrol.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6HK3
 
 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C44 | | Descriptor: | 3-azanyl-~{N}-(2-methoxyphenyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
7CJ3
 
 | | Crystal structure of the transmembrane domain of Salpingoeca rosetta rhodopsin phosphodiesterase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphodiesterase, RETINAL | | Authors: | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
