6OGO
 
 | | Crystal structure of NDM-9 metallo-beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
8TXS
 
 | |
8TUP
 
 | | Cryo-EM structure of the human MRS2 magnesium channel under Mg2+-free condition | | Descriptor: | MAGNESIUM ION, Magnesium transporter MRS2 homolog, mitochondrial | | Authors: | Lai, L.T.F, Balaraman, J, Zhou, F, Matthies, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human magnesium channel MRS2 reveal gating and regulatory mechanisms.
Nat Commun, 14, 2023
|
|
6HZC
 
 | |
7QLJ
 
 | |
8TUL
 
 | | Cryo-EM structure of the human MRS2 magnesium channel under Mg2+ condition | | Descriptor: | MAGNESIUM ION, Magnesium transporter MRS2 homolog, mitochondrial | | Authors: | Lai, L.T.F, Balaraman, J, Zhou, F, Matthies, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human magnesium channel MRS2 reveal gating and regulatory mechanisms.
Nat Commun, 14, 2023
|
|
6OL5
 
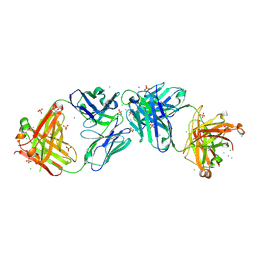 | |
8PIW
 
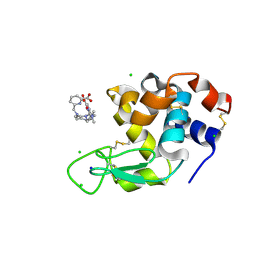 | | Crystal structure of Hen Egg White Lysozyme co-crystallized with 10 mM TbXo4-NMet2 | | Descriptor: | 6-[[4-[(6-carboxypyridin-2-yl)methyl]-7-[3-(dimethylamino)propyl]-1,4,7-triazonan-1-yl]methyl]pyridine-2-carboxylic acid, CHLORIDE ION, Lysozyme C, ... | | Authors: | Alsalman, Z, Girard, E. | | Deposit date: | 2023-06-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Influence of Chemical Modifications of the Crystallophore on Protein Nucleating Properties and Supramolecular Interactions Network.
Chemistry, 30, 2024
|
|
6IBZ
 
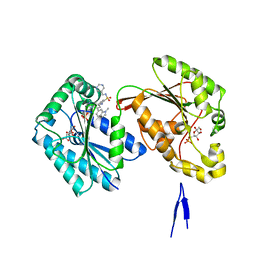 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 7 | | Descriptor: | 2-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-(pyrimidin-5-ylmethyl)benzenesulfonamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Tomczyk, M, Guzik, P, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6RHJ
 
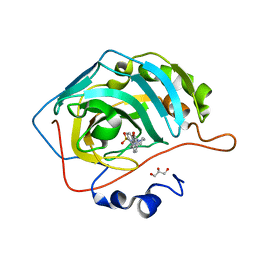 | | Crystal structure of human carbonic anhydrase isozyme II with 4-(4-benzyl-1,4-diazepane-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-[[4-(phenylmethyl)-1,4-diazepan-1-yl]carbonyl]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
7QTB
 
 | | S1 nuclease from Aspergillus oryzae in complex with cytidine-5'-monophosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Atomic resolution studies of S1 nuclease complexes reveal details of RNA interaction with the enzyme despite multiple lattice-translocation defects.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6RG4
 
 | | Crystal structure of human Carbonic anhydrase II in complex with (R)-4-(2-benzyl-4-methylpiperazin-1-yl)benzenesulfonamide | | Descriptor: | 4-[(3~{S})-4-methyl-3-(phenylmethyl)piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
7QTA
 
 | | S1 nuclease from Aspergillus oryzae in complex with uridine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nuclease S1, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic resolution studies of S1 nuclease complexes reveal details of RNA interaction with the enzyme despite multiple lattice-translocation defects.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6OL6
 
 | | Structure of iglb12 scFv in complex with anti-idiotype ib2 Fab | | Descriptor: | CHLORIDE ION, ib2 Heavy Chain, ib2 Light Chain, ... | | Authors: | Weidle, C, Pancera, M, Gewe, M. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Detection and activation of HIV broadly neutralizing antibody precursor B cells using anti-idiotypes.
J.Exp.Med., 216, 2019
|
|
8TNK
 
 | |
6O5J
 
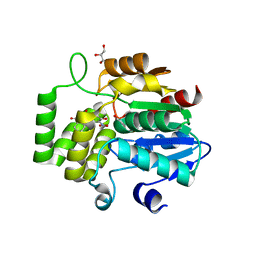 | | Crystal Structure of DAD2 bound to quinazolinone derivative | | Descriptor: | 1-(4-hydroxy-3-nitrophenyl)quinazoline-2,4(1H,3H)-dione, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hamiaux, C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Chemical synthesis and characterization of a new quinazolinedione competitive antagonist for strigolactone receptors with an unexpected binding mode.
Biochem.J., 476, 2019
|
|
6RJ1
 
 | |
5MFW
 
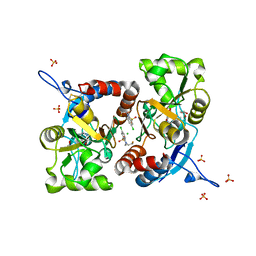 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-121 at 2.10 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | Authors: | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
6RIV
 
 | | Crystal structure of Alopecurus myosuroides GSTF | | Descriptor: | GLYCEROL, Glutathione transferase, S-Hydroxy-Glutathione, ... | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2019-04-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Comparative structural and functional analysis of phi class glutathione transferases involved in multiple-herbicide resistance of grass weeds and crops.
Plant Physiol Biochem., 149, 2020
|
|
6RT9
 
 | |
7QB2
 
 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7QFM
 
 | | Pim1 in complex with (E)-4-((2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
6RTA
 
 | |
8TN3
 
 | |
7QG0
 
 | | Inhibitor-induced hSARM1 duplex | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Zalk, R, Kahzma, T, Guez-Haddad, J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A duplex structure of SARM1 octamers stabilized by a new inhibitor.
Cell.Mol.Life Sci., 80, 2022
|
|
