4IBT
 
 | | Human p53 core domain with hot spot mutation R273H and second-site suppressor mutation T284R | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Diskin-Posner, Y, Eldar, A, Rozenberg, H, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
4IE4
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with 5-carboxy-8-hydroxyquinoline (IOX1) | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5048 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
3F5Q
 
 | | CRYSTAL STRUCTURE OF putative short chain dehydrogenase from Escherichia coli CFT073 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
4IHG
 
 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
4ILU
 
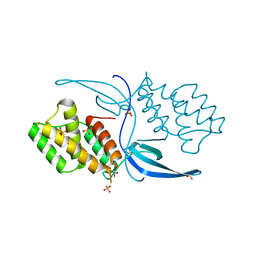 | | Crystal structure of Mycobacterium tuberculosis CarD | | Descriptor: | RNA polymerase-binding transcription factor CarD, SULFATE ION | | Authors: | Thakur, K.G, Kaur, G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis CarD, an essential RNA polymerase binding protein, reveals a quasidomain-swapped dimeric structural architecture.
Proteins, 82, 2014
|
|
3FMW
 
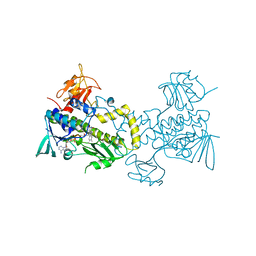 | | The crystal structure of MtmOIV, a Baeyer-Villiger monooxygenase from the mithramycin biosynthetic pathway in Streptomyces argillaceus. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase | | Authors: | Noinaj, N, Beam, M.P, Wang, C, Rohr, J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of Baeyer-Villiger monooxygenase MtmOIV, the key enzyme of the mithramycin biosynthetic pathway .
Biochemistry, 48, 2009
|
|
3FOP
 
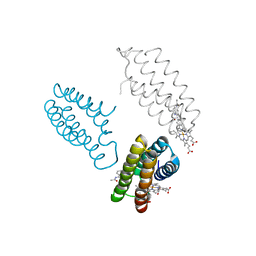 | |
4IQA
 
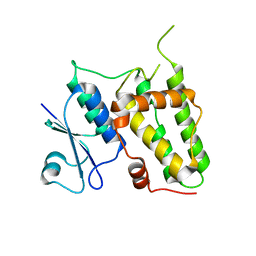 | |
3FH6
 
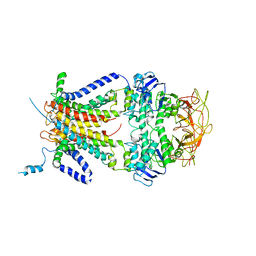 | | Crystal structure of the resting state maltose transporter from E. coli | | Descriptor: | Maltose transport system permease protein malF, Maltose transport system permease protein malG, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Khare, D, Oldham, M.L, Orelle, C, Davidson, A.L, Chen, J. | | Deposit date: | 2008-12-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Alternating access in maltose transporter mediated by rigid-body rotations.
Mol.Cell, 33, 2009
|
|
3FPG
 
 | | Crystal Structure of E81Q mutant of MtNAS | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, Putative uncharacterized protein | | Authors: | Dreyfus, C, Pignol, D, Arnoux, P. | | Deposit date: | 2009-01-05 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshots of iterative substrate translocations during nicotianamine synthesis in Archaea
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FLP
 
 | |
4H9B
 
 | | Radiation damage study of lysozyme - 0.70 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.1998 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3F8M
 
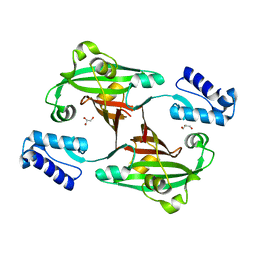 | | Crystal Structure of PhnF from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, GntR-family protein transcriptional regulator | | Authors: | Busby, J.N, Gebhard, S, Cook, G.M, Lott, S.J, Baker, E.N, Money, V.A. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PhnF, a GntR-family transcription regulator in Mycobacterium smegmatis
To be Published
|
|
4H9H
 
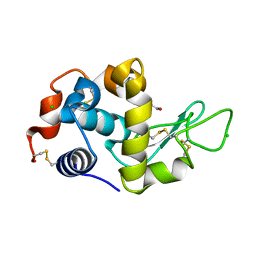 | | Radiation damage study of lysozyme - 0.98 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.2002 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H98
 
 | | Candida glabrata dihydrofolate reductase complexed with NADPH and 5-{3-[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine (UCP1018) | | Descriptor: | 5-{3-[3-(1,3-benzodioxol-5-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine, Dihydrofolate Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-09-24 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the active sites of dihydrofolate reductase from two species of Candida uncovers ligand-induced conformational changes shared among species.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HBC
 
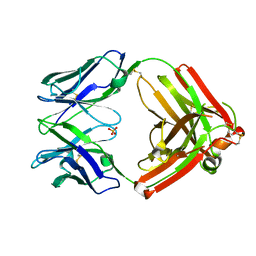 | |
3FN8
 
 | |
3FNM
 
 | | Crystal structure of acivicin-inhibited gamma-glutamyltranspeptidase reveals critical roles for its C-terminus in autoprocessing and catalysis | | Descriptor: | (2S)-AMINO[(5S)-3-CHLORO-4,5-DIHYDROISOXAZOL-5-YL]ACETIC ACID, Gamma-glutamyltranspeptidase (Ggt) Large subunit, Gamma-glutamyltranspeptidase (Ggt) Small subunit | | Authors: | Williams, K, Cullati, S, Sand, A, Biterova, E.I, Barycki, J.J. | | Deposit date: | 2008-12-25 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Acivicin-Inhibited gamma-Glutamyltranspeptidase Reveals Critical Roles for Its C-Terminus in Autoprocessing and Catalysis.
Biochemistry, 48, 2009
|
|
4HBW
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-ethyl-3-methyl-2-oxo-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4HE0
 
 | | Crystal structure of human muscle fructose-1,6-bisphosphatase | | Descriptor: | CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, MAGNESIUM ION, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
3FMD
 
 | | Crystal Structure of Human Haspin with an Isoquinoline ligand | | Descriptor: | 1,2-ETHANEDIOL, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, NICKEL (II) ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Haspin with an Isoquinoline ligand
To be Published
|
|
4GPJ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a isoxazolylbenzimidazole ligand | | Descriptor: | (1R)-6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-phenyl-2,3-dihydro-1H-inden-1-ol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Heightman, T.D, Brennan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The design and synthesis of 5- and 6-isoxazolylbenzimidazoles as selective inhibitors of the BET bromodomains.
Medchemcomm, 4, 2013
|
|
4GRS
 
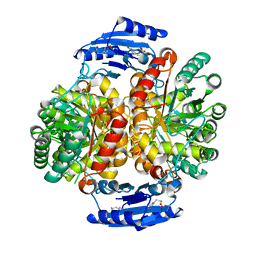 | | Crystal structure of a chimeric DAH7PS | | Descriptor: | Phospho-2-dehydro-3-deoxyheptonate aldolase, 2-dehydro-3-deoxyphosphoheptonate aldolase, TYROSINE | | Authors: | Cross, P.J, Allison, T.M, Dobson, R.C.J, Jameson, G.B, Parker, E.J. | | Deposit date: | 2012-08-26 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering allosteric control to an unregulated enzyme by transfer of a regulatory domain
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GOS
 
 | | Crystal structure of human B7-H4 IgV-like domain | | Descriptor: | V-set domain-containing T-cell activation inhibitor 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Vigdorovich, V, Ramagopal, U, Bhosle, R, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and cancer immunotherapy of the B7 family member B7x.
Cell Rep, 9, 2014
|
|
4GSB
 
 | |
