8BU7
 
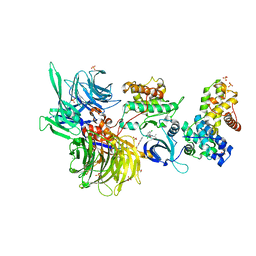 | | Structure of DDB1 bound to 21195-engaged CDK12-cyclin K | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,6-bis(chloranyl)phenyl]-6-[[4-(2-hydroxyethyloxy)phenyl]methyl]-3-propan-2-yl-5H-pyrazolo[3,4-d]pyrimidin-4-one, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
7P0U
 
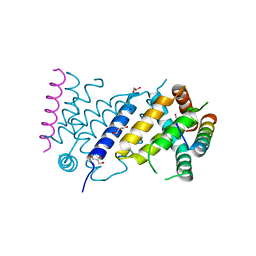 | | ORF virus encoded Bcl-2 homolog ORFV125 in complex with Puma BH3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Activator of apoptosis harakiri, Apoptosis inhibitor, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99374259 Å) | | Cite: | Structural Investigation of Orf Virus Bcl-2 Homolog ORFV125 Interactions with BH3-Motifs from BH3-Only Proteins Puma and Hrk.
Viruses, 13, 2021
|
|
9NIJ
 
 | |
9NIM
 
 | |
6YY8
 
 | | Crystal structure of SAICAR Synthetase (PurC) from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[1-(2-morpholin-4-ylethyl)pyrazol-4-yl]pyrimidine-5-carbonitrile, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Thomas, S.E, Charoensutthivarakul, S, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2020-05-04 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Development of Inhibitors of SAICAR Synthetase (PurC) from Mycobacterium abscessus Using a Fragment-Based Approach.
Acs Infect Dis., 8, 2022
|
|
9NIK
 
 | |
6ON1
 
 | | A resting state structure of L-DOPA dioxygenase from Streptomyces sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, L-DOPA dioxygenase | | Authors: | Wang, Y, Shin, I, Fu, Y, Colabroy, K, Liu, A. | | Deposit date: | 2019-04-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Crystal Structures of L-DOPA Dioxygenase fromStreptomyces sclerotialus.
Biochemistry, 58, 2019
|
|
9FJF
 
 | | Lysosomal transporting complex of beta-glucocerebrosidase (GCase) and lysosomal integral membrane protein 2 (LIMP-2) with bound Pro-macrobodies (Combined focus map) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dobert, J.P, Schaefer, J.H.S, Dal Maso, T, Socher, E, Versees, W, Moeller, A, Zunke, F, Arnold, P. | | Deposit date: | 2024-05-31 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-TEM structure of beta-glucocerebrosidase in complex with its transporter LIMP-2.
Nat Commun, 16, 2025
|
|
8BU3
 
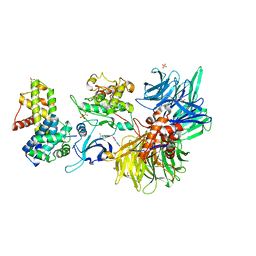 | | Structure of DDB1 bound to DS19-engaged CDK12-cyclin K | | Descriptor: | 1,2-ETHANEDIOL, 2-morpholin-4-yl-9-propan-2-yl-~{N}-[(4-pyridin-2-ylphenyl)methyl]purin-6-amine, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
6OOZ
 
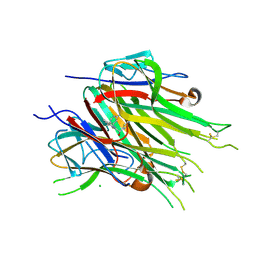 | | Asymmetric hTNF-alpha | | Descriptor: | (S)-{1-[(2,5-dimethylphenyl)methyl]-1H-benzimidazol-2-yl}(pyridin-4-yl)methanol, CHLORIDE ION, Tumor necrosis factor | | Authors: | Arakaki, T.L, Edwards, T.E, Fox III, D, Lecomte, F, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
8BUS
 
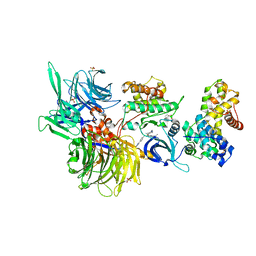 | | Structure of DDB1 bound to DS59-engaged CDK12-cyclin K | | Descriptor: | 1,3-dimethyl-5-[[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)methylamino]purin-2-yl]amino]methyl]pyrazole-4-sulfonamide, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
6XJ8
 
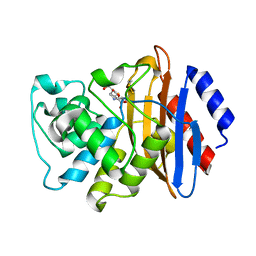 | | KPC-2 N170A mutant bound to hydrolyzed imipenem at 2.05 A | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2020-06-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | KPC-2 beta-lactamase enables carbapenem antibiotic resistance through fast deacylation of the covalent intermediate.
J.Biol.Chem., 296, 2020
|
|
7NI4
 
 | | Human ATM kinase domain with bound M4076 inhibitor | | Descriptor: | 8-(1,3-dimethylpyrazol-4-yl)-1-(3-fluoranyl-5-methoxy-pyridin-4-yl)-7-methoxy-3-methyl-imidazo[4,5-c]quinolin-2-one, Serine-protein kinase ATM, ZINC ION | | Authors: | Stakyte, K, Rotheneder, M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of human ATM kinase inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5MRI
 
 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with Triazolone 18 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2016-12-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The identification of novel acid isostere based inhibitors of the VPS10P family sorting receptor Sortilin.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
9NNF
 
 | | Cryo-EM structure of a de-novo designed binder NY1-B04 in complex with HLA-A*02:01 and NY-ESO-1-derived peptide SLLMWITQC | | Descriptor: | Beta-2-microglobulin, De-novo designed binder NY1-B04, HLA class I histocompatibility antigen, ... | | Authors: | Gharpure, A, Fernandez-Quintero, M.L, Ward, A.B. | | Deposit date: | 2025-03-05 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | De novo-designed pMHC binders facilitate T cell-mediated cytotoxicity toward cancer cells.
Science, 389, 2025
|
|
9OUG
 
 | | Influenza A Virus Nucleoprotein(8-498)NP complex with rac-5-(4-(((2R,6R)-6-(methoxymethyl)-6-methyl-1,4-dioxan-2-yl)methoxy)phenyl)-2-oxo-6-(trifluoromethyl)-1,2-dihydropyridine-3-carboxamide (Compound 20) | | Descriptor: | 5-(4-{[(2R,6S)-6-(methoxymethyl)-6-methyl-1,4-dioxan-2-yl]methoxy}phenyl)-2-oxo-6-(trifluoromethyl)-1,2-dihydropyridine-3-carboxamide, Nucleoprotein | | Authors: | Mamo, M. | | Deposit date: | 2025-05-28 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery and Optimization of a Novel Series of Influenza A Virus Replication Inhibitors Targeting the Nucleoprotein Protein-Protein Interaction.
J.Med.Chem., 68, 2025
|
|
7P85
 
 | |
6NDL
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with a sulfonamide inhibitor | | Descriptor: | 1-[4-(6-aminopurin-9-yl)butylsulfamoyl]-3-[4-[(4~{S})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butyl]urea, Biotin Protein Ligase, GLYCEROL | | Authors: | Marshall, A.C, Polyak, S.W, Bruning, J.B, Lee, K. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads.
Acs Chem.Biol., 14, 2019
|
|
7N6T
 
 | |
5MUJ
 
 | | BT0996 RGII Chain B Complex | | Descriptor: | ACETATE ION, Beta-galactosidase, alpha-L-rhamnopyranose-(1-2)-[alpha-L-rhamnopyranose-(1-3)]alpha-L-arabinopyranose-(1-4)-[4-O-[(1R)-1-hydroxyethyl]-2-O-methyl-alpha-L-fucopyranose-(1-2)]beta-D-galactopyranose-(1-2)-alpha-D-aceric acid-(1-4)-alpha-L-rhamnopyranose-(1-3)-3-C-(hydroxylmethyl)-alpha-D-erythrofuranose | | Authors: | Cartmell, A, Basle, A, Ndeh, D, Luis, A.S, Venditto, I, Labourel, A, Rogowski, A, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
6Z9V
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting IIGWMWIPV | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
7NG7
 
 | | Src kinase bound to eCF506 trapped in inactive conformation | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase Src, tert-butyl (4-(4-amino-1-(2-(4-(dimethylamino)piperidin-1-yl)ethyl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl)-2-methoxyphenyl)carbamate | | Authors: | Lietha, D, Unciti-Broceta, A. | | Deposit date: | 2021-02-08 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Conformation Selective Mode of Inhibiting SRC Improves Drug Efficacy and Tolerability.
Cancer Res., 81, 2021
|
|
5MYB
 
 | | Homodimerization of Tie2 Fibronectin-like domains 2 and 3 in space group P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiopoietin-1 receptor, ... | | Authors: | Leppanen, V.-M, Saharinen, P, Alitalo, K. | | Deposit date: | 2017-01-26 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of Tie2 activation and Tie2/Tie1 heterodimerization.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6Z9Z
 
 | | Atomic resolution X-ray structure of the Uridine phosphorylase from Vibrio cholerae on crystals grown under microgravity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Eistrikh-Heller, P.A, Rubinsky, S.V, Samygina, V.R, Gabdulkhakov, A.G, Lashkov, A.A. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystallization in Microgravity and the Atomic-Resolution Structure of Uridine Phosphorylase from Vibrio cholerae
Crystallography Reports, 66, 2021
|
|
7P9Y
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
