8U72
 
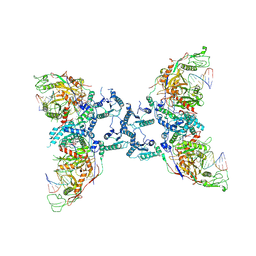 | | Cryo-EM structure of the SPARTA oligomer with guide RNA and target DNA | | Descriptor: | DNA (5'-D(P*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*C)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Malik, R, Kottur, J, Aggarwal, A.K. | | Deposit date: | 2023-09-14 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Nucleic acid mediated activation of a short prokaryotic Argonaute immune system.
Nat Commun, 15, 2024
|
|
3LGB
 
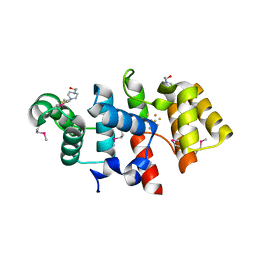 | |
4IX9
 
 | | Crystal structure of subunit F of V-ATPase from S. cerevisiae | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, V-type proton ATPase subunit F | | Authors: | Basak, S, Balakrishna, A.M, Manimekalai, M.S.S, Gruber, G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal and NMR structures give insights into the role and dynamics of subunit F of the eukaryotic V-ATPase from Saccharomyces cerevisiae
J.Biol.Chem., 288, 2013
|
|
4F2N
 
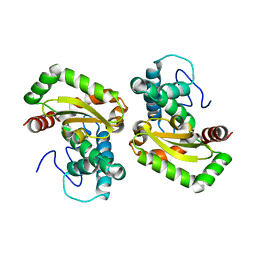 | |
4GG2
 
 | | The crystal structure of glutamate-bound human gamma-glutamyltranspeptidase 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-04 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
5MK8
 
 | |
5M9S
 
 | | Human angiogenin ALS variant V103I | | Descriptor: | Angiogenin, D(-)-TARTARIC ACID | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
5MTK
 
 | | Crystal structure of human Caspase-1 with (3S,6S,10aS)-N-((2S,3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-6-(isoquinoline-1-carboxamido)-5-oxodecahydropyrrolo[1,2-a]azocine-3-carboxamide (PGE-3935199) | | Descriptor: | (3~{S})-3-[[(3~{S},6~{S},10~{a}~{S})-6-(isoquinolin-1-ylcarbonylamino)-5-oxidanylidene-2,3,6,7,8,9,10,10~{a}-octahydro-1~{H}-pyrrolo[1,2-a]azocin-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2017-01-09 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Playing against the odds: scaffold hopping from 3D-fragments
To Be Published
|
|
4RDL
 
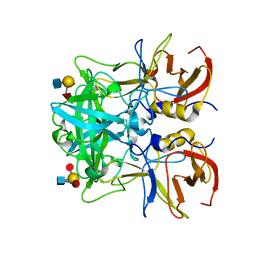 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
4RDK
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis b tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
5MK7
 
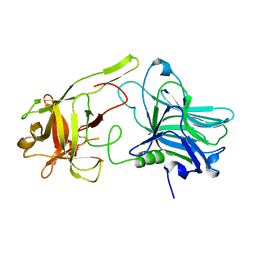 | |
5M9V
 
 | | Human angiogenin PD/ALS variant R121C | | Descriptor: | Angiogenin, GLUTATHIONE, L(+)-TARTARIC ACID | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
5MWC
 
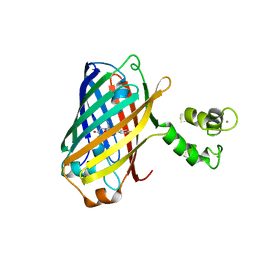 | | Crystal structure of the genetically-encoded green calcium indicator NTnC in its calcium bound state | | Descriptor: | CALCIUM ION, genetically-encoded green calcium indicator NTnC | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Rakitina, T.V, Popov, V.O, Subach, O.M, Barykina, N.V, Subach, F.V. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enchanced variant of genetically-encoded green calcium indicator NTnC
To Be Published
|
|
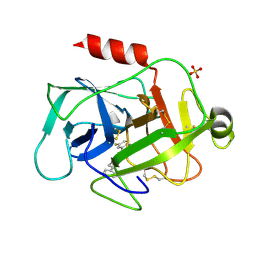
5MT0
 
 | |
5MMV
 
 | | Crystal structure of human Caspase-1 with 2-((2-naphthoyl)-L-valyl)-4-hydroxy-N-((3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-2-azabicyclo[2.2.2]octane-3-carboxamide (Compound 1) | | Descriptor: | (3~{S})-3-[[(3~{S})-2-[(2~{S})-3-methyl-2-(naphthalen-2-ylcarbonylamino)butanoyl]-4-oxidanyl-2-azabicyclo[2.2.2]octan-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human Caspase-1 with 2-((2-naphthoyl)-L-valyl)-4-hydroxy-N-((3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-2-azabicyclo[2.2.2]octane-3-carboxamide (Compound 1)
To Be Published
|
|
4RAK
 
 | |
5MT4
 
 | |
4RA8
 
 | | Structure analysis of the Mip1a P8A mutant | | Descriptor: | C-C motif chemokine 3 | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4RAL
 
 | | Crystal structure of insulin degrading enzyme in complex with macrophage inflammatory protein 1 beta | | Descriptor: | C-C motif chemokine 4, Insulin-degrading enzyme, ZINC ION | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
8TWX
 
 | |
8V7H
 
 | |
8V7E
 
 | | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mg2+
To Be Published
|
|
8V7A
 
 | |
8V7C
 
 | |
8V7K
 
 | |
