5AAA
 
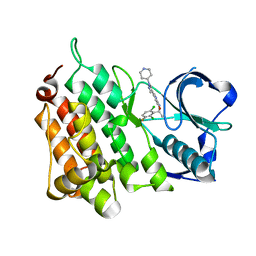 | | Structure of L1198F Mutant Human Anaplastic Lymphoma Kinase in Complex with Crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Resensitization to Crizotinib by the Lorlatinib Alk Resistance Mutation L1198F.
N.Engl.J.Med., 374, 2016
|
|
7XXD
 
 | | Orf1-sarcosine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-formimidoyl fortimicin A synthase, SARCOSINE | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7Y0X
 
 | | Orf1-glycine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-06-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXR
 
 | | Orf1 R342A-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-formimidoyl fortimicin A synthase, [(2~{R},3~{R},4~{S},5~{R},6~{R})-6-[(~{E})-[(3~{a}~{S},7~{R},7~{a}~{S})-7-oxidanyl-4-oxidanylidene-3,3~{a},5,6,7,7~{a}-hexahydro-1~{H}-imidazo[4,5-c]pyridin-2-ylidene]amino]-5-(2-azanylethanoylamino)-2-(hydroxymethyl)-4-oxidanyl-oxan-3-yl] carbamate | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
4ZY1
 
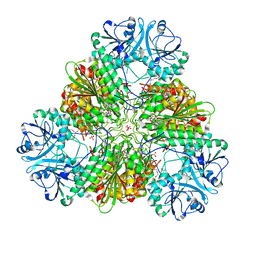 | |
7XXP
 
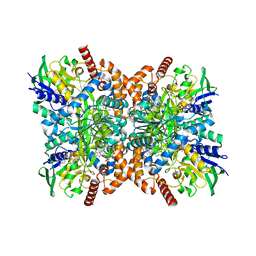 | | F316A-glycine-streptothricin F complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
5DPN
 
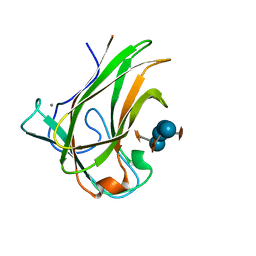 | | Engineered CBM X-2 L110F in complex with branched carbohydrate XXXG. | | Descriptor: | CALCIUM ION, Xylanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ohlin, M. | | Deposit date: | 2015-09-13 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Neutron Crystallographic Studies Reveal Hydrogen Bond and Water-Mediated Interactions between a Carbohydrate-Binding Module and Its Bound Carbohydrate Ligand.
Biochemistry, 54, 2015
|
|
5DRX
 
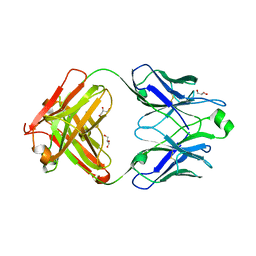 | |
3LRS
 
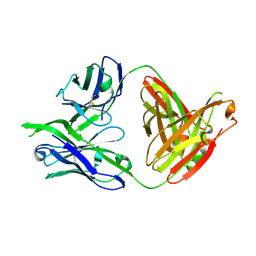 | | Structure of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PG-16 Heavy Chain Fab, PG-16 Light Chain Fab | | Authors: | Pancera, M, Kwong, P.D. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
5DRW
 
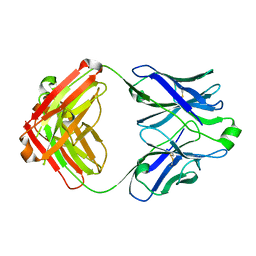 | |
5AYF
 
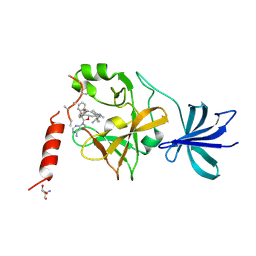 | | Crystal structure of SET7/9 in complex with cyproheptadine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(dibenzo[1,2-a:2',1'-d][7]annulen-11-ylidene)-1-methyl-piperidine, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Niwa, H, Handa, N, Takemoto, Y, Ito, A, Tomabechi, Y, Umehara, T, Shirouzu, M, Yoshida, M, Yokoyama, S. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Identification of Cyproheptadine as an Inhibitor of SET Domain Containing Lysine Methyltransferase 7/9 (Set7/9) That Regulates Estrogen-Dependent Transcription
J.Med.Chem., 59, 2016
|
|
5B1M
 
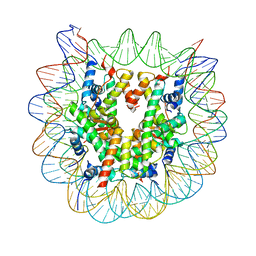 | | The mouse nucleosome structure containing H3.1 | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B type 3-A, ... | | Authors: | Urahama, T, Machida, S, Horikoshi, N, Osakabe, A, Tachiwana, H, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2015-12-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Testis-Specific Histone Variant H3t Gene Is Essential for Entry into Spermatogenesis
Cell Rep, 18, 2017
|
|
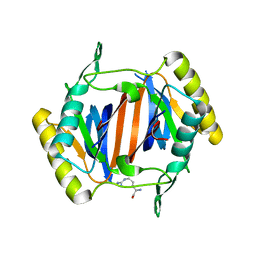
3KO9
 
 | |
5AX9
 
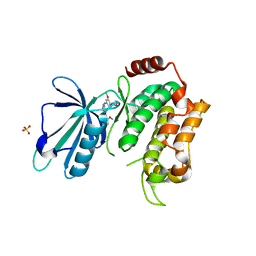 | | Crystal structure of the kinase domain of human TRAF2 and NCK-interacting protein kinase in complex with compund 9 | | Descriptor: | 4-methoxy-3-[2-[(3-methoxy-4-morpholin-4-yl-phenyl)amino]pyridin-4-yl]benzenecarbonitrile, SULFATE ION, TRAF2 and NCK-interacting protein kinase | | Authors: | Ohbayashi, N, Kukimoto-Niino, M, Yamada, T, Shirouzu, M. | | Deposit date: | 2015-07-21 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | TNIK inhibition abrogates colorectal cancer stemness
Nat Commun, 7, 2016
|
|
5B1L
 
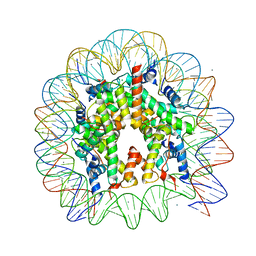 | | The mouse nucleosome structure containing H3t | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1, ... | | Authors: | Urahama, T, Machida, S, Horikoshi, N, Osakabe, A, Tachiwana, H, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2015-12-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Testis-Specific Histone Variant H3t Gene Is Essential for Entry into Spermatogenesis
Cell Rep, 18, 2017
|
|
3KOB
 
 | |
7XAR
 
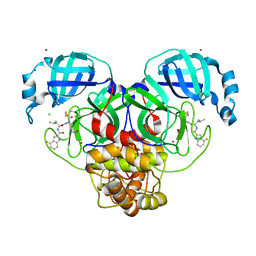 | | Crystal structure of 3C-like protease from SARS-CoV-2 in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase, 4-fluoranyl-~{N}-[(2~{S})-1-[2-(2-fluoranylethanoyl)-2-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]hydrazinyl]-4-methyl-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Ochi, J, Takahashi, D, Ueda, T, Ojida, A. | | Deposit date: | 2022-03-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease.
J.Med.Chem., 65, 2022
|
|
3KO4
 
 | |
3KOC
 
 | |
3KNF
 
 | |
7YZV
 
 | |
7Z09
 
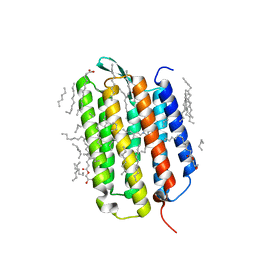 | | Crystal structure of the ground state of bacteriorhodopsin at 1.05 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0A
 
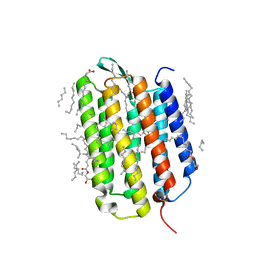 | | Crystal structure of the ground state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0D
 
 | | Crystal structure of the L state of bacteriorhodopsin at 1.20 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0C
 
 | | Crystal structure of the K state of bacteriorhodopsin at 1.53 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
