4HXS
 
 | | Brd4 Bromodomain 1 complex with N-[3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)PHENYL]-1-PHENYLMETHANESULFONAMIDE inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]-1-phenylmethanesulfonamide | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
8FML
 
 | |
4HYO
 
 | | Crystal Structure of MthK Pore | | Descriptor: | Calcium-gated potassium channel mthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, McCoy, J.G, Nimigean, C.M. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The voltage-dependent gate in MthK potassium channels is located at the selectivity filter.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4HVW
 
 | |
4HZK
 
 | |
4I1U
 
 | |
4HWR
 
 | |
4IME
 
 | |
4IBJ
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(5S)-3-hydroxy-2-oxo-4-[3-(trifluoromethyl)benzoyl]-5-[3-(trifluoromethyl)phenyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IOY
 
 | |
4IE1
 
 | | Crystal structure of human Arginase-1 complexed with inhibitor 1h | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-amino-5-carboxy-8-hydroxyoctyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Beckett, P, Van Zandt, M.C, Ji, M.K, Whitehouse, D, Ryder, T, Jagdmann, E, Andreoli, M, Mazur, A, Padmanilayam, M, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | 2-Substituted-2-amino-6-boronohexanoic acids as arginase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IHF
 
 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | Acyl carrier protein, CHLORIDE ION, S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] (3R)-3-hydroxytetradecanethioate, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
4IJN
 
 | |
4IBI
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-4-hydroxy-5-oxo-3-[3-(trifluoromethyl)benzoyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IKN
 
 | | Crystal structure of adaptor protein complex 3 (AP-3) mu3A subunit C-terminal domain, in complex with a sorting peptide from TGN38 | | Descriptor: | AP-3 complex subunit mu-1, Trans-Golgi network integral membrane protein TGN38 | | Authors: | Mardones, G.A, Kloer, D.P, Burgos, P.V, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2012-12-26 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for the recognition of tyrosine-based sorting signals by the mu 3A subunit of the AP-3 adaptor complex.
J.Biol.Chem., 288, 2013
|
|
4IM4
 
 | |
4IH8
 
 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, Calmodulin-domain protein kinase 1 | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, K, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.877 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
To be Published
|
|
4IJ0
 
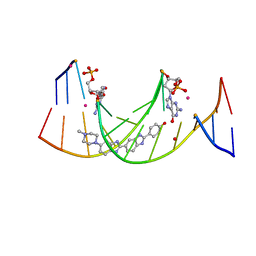 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), STRONTIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
4IOO
 
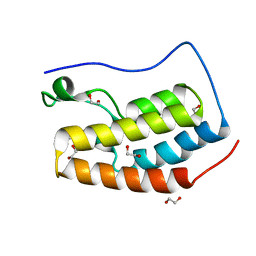 | | Crystal Structure of the first bromodomain of BRD4 in complex with N-methyltrimethylacetamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Lolli, G, Battistutta, R. | | Deposit date: | 2013-01-08 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Different orientations of low-molecular-weight fragments in the binding pocket of a BRD4 bromodomain.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IBE
 
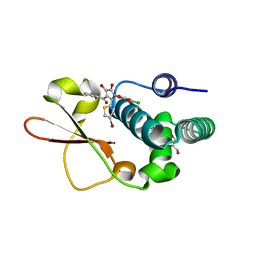 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-chlorobenzoic acid, GLYCEROL, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBV
 
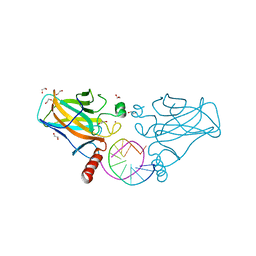 | | Human p53 core domain with hot spot mutation R273C and second-site suppressor mutation S240R in sequence-specific complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA (5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3'), ... | | Authors: | Eldar, A, Rozenberg, H, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
3COQ
 
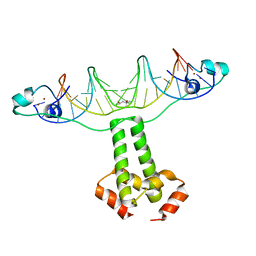 | | Structural Basis for Dimerization in DNA Recognition by Gal4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*DAP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DAP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DTP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), ... | | Authors: | Hong, M, Fitzgerald, M.X, Harper, S, Luo, C, Speicher, D.W. | | Deposit date: | 2008-03-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dimerization in DNA recognition by gal4.
Structure, 16, 2008
|
|
4IBF
 
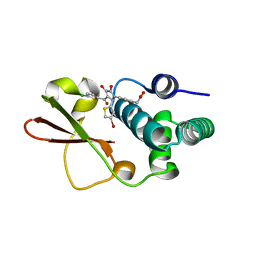 | | Ebola virus VP35 bound to small molecule | | Descriptor: | (4-{(2R)-2-(4-bromothiophen-2-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}phenyl)acetic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4ID1
 
 | |
4IJ8
 
 | | Crystal structure of the complex of SETD8 with SAM | | Descriptor: | N-lysine methyltransferase SETD8, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION, ... | | Authors: | Yu, W, Tempel, W, Li, Y, El Bakkouri, M, Shapira, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of SETD8 with SAM
To be Published
|
|
