6BCJ
 
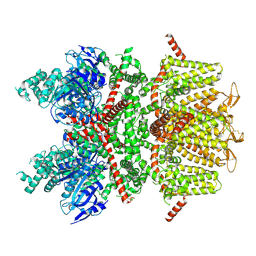 | | cryo-EM structure of TRPM4 in apo state with short coiled coil at 3.1 angstrom resolution | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
6SIM
 
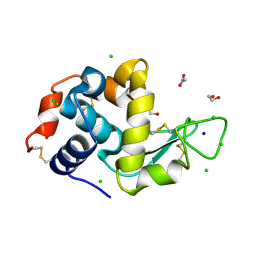 | | SAD structure of Hen Egg White Lysozyme recovered by inverse beam geometry data collection and univariate analysis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Garcia-Bonete, M.J, Katona, G. | | Deposit date: | 2019-08-10 | | Release date: | 2019-11-06 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.61018026 Å) | | Cite: | Bayesian machine learning improves single-wavelength anomalous diffraction phasing.
Acta Crystallogr.,Sect.A, 75, 2019
|
|
5FU3
 
 | | The complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition | | Descriptor: | CBM74-RFGH5, SODIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Basle, A, Luis, A.S, Venditto, I, Gilbert, H.J. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6BG0
 
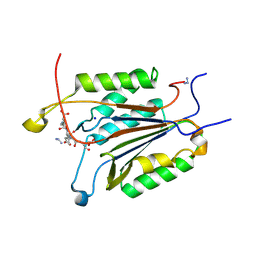 | | Caspase-3 Mutant - D9A,D28A,S150D | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, AZIDE ION, Caspase-3, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-27 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.125 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6AWN
 
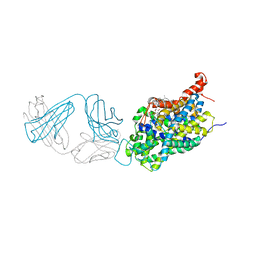 | |
6AWZ
 
 | | Structure of PR-10 allergen from peanut (Ara h 8.01). | | Descriptor: | Ara h 8 allergen, SODIUM ION, TETRAETHYLENE GLYCOL | | Authors: | Offermann, L.R, Majorek, K, McBride, J, Hurlburt, B.K, Maleki, S.J, Pote, S.S, Chruszcz, M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of PR-10 Allergen Ara h 8.01.
To Be Published
|
|
6B2X
 
 | | Apo YiuA Crystal Form 1 | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute-binding periplasmic protein of iron/siderophore ABC transporter | | Authors: | Radka, C.D, DeLucas, L.J, Aller, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The crystal structure of the Yersinia pestis iron chaperone YiuA reveals a basic triad binding motif for the chelated metal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6SXE
 
 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with Endoxifen (2.6 Angstrom resolution) | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, Endoxifen, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
4LS8
 
 | | Crystal structure of Bacillus subtilis beta-ketoacyl-ACP synthase II (FabF) in a covalent complex with cerulenin | | Descriptor: | (3R,7E,10E)-3-hydroxy-4-oxododeca-7,10-dienamide, 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-07-22 | | Release date: | 2014-04-02 | | Last modified: | 2014-06-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into bacterial resistance to cerulenin.
Febs J., 281, 2014
|
|
6B9I
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with 14-Methylhexadecanoic Acid (Anteiso C17:0) to 1.93 Angstrom resolution | | Descriptor: | (14S)-14-methylhexadecanoic acid, Fatty acid Kinase (Fak) B1, GLYCEROL, ... | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-10-10 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Acyl-chain selectivity and physiological roles ofStaphylococcus aureusfatty acid-binding proteins.
J. Biol. Chem., 294, 2019
|
|
6SX5
 
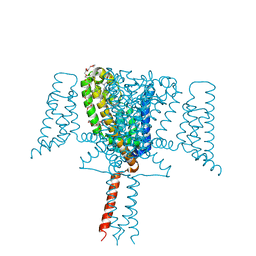 | | Full length Mutant Open-form Sodium Channel NavMs | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
3B35
 
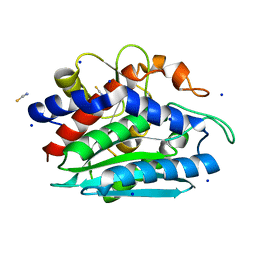 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus | | Descriptor: | Bacterial leucyl aminopeptidase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-19 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
6AWO
 
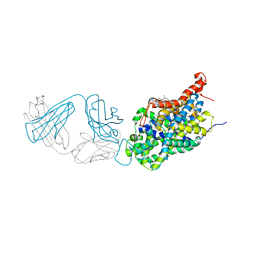 | | X-ray structure of the ts3 human serotonin transporter complexed with sertraline at the central site | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody FAB heavy chain, ... | | Authors: | Coleman, J.A, Gouaux, E. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.534 Å) | | Cite: | Structural basis for recognition of diverse antidepressants by the human serotonin transporter.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6AWP
 
 | |
5WNY
 
 | | DNA polymerase beta substrate complex with incoming 5-FdUTP | | Descriptor: | 2'-deoxy-5-fluorouridine 5'-(tetrahydrogen triphosphate), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
5FQU
 
 | | Orthorhombic crystal structure of of PlpD (selenomethionine derivative) | | Descriptor: | AZIDE ION, PATATIN-LIKE PROTEIN, PLPD, ... | | Authors: | Vinicius da Mata Madeira, P, Zouhir, S, Basso, P, Neves, D, Laubier, A, Salacha, R, Bleves, S, Faudry, E, Contreras-Martel, C, Dessen, A. | | Deposit date: | 2015-12-14 | | Release date: | 2016-04-06 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis of Lipid Targeting and Destruction by the Type V Secretion System of Pseudomonas Aeruginosa.
J.Mol.Biol., 428, 2016
|
|
6SJ8
 
 | | Methyltransferase MtgA from Desulfitobacterium hafniense in complex with tetrahydrofolate | | Descriptor: | (6S)-2-amino-6-methyl-5,6,7,8-tetrahydropteridin-4(3H)-one, GLYCEROL, SODIUM ION, ... | | Authors: | Badmann, T, Groll, M. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures in Tetrahydrofolate Methylation in Desulfitobacterial Glycine Betaine Metabolism at Atomic Resolution.
Chembiochem, 21, 2020
|
|
5FUB
 
 | | Crystal Structure of zebrafish Protein Arginine Methyltransferase 2 catalytic domain with SAH | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-01-22 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
6B4N
 
 | | a hydroxymethyl functionality at the 4-position of the 2-phenyloxazole moiety of HIV-1 protease inhibitors involving the P2' ligands | | Descriptor: | CHLORIDE ION, Protease, SODIUM ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and X-ray Studies of HIV-1 Protease Inhibitors with Modified P2' Ligands of Darunavir.
ChemMedChem, 12, 2017
|
|
6SUY
 
 | |
5W8S
 
 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations | | Descriptor: | LITHIUM ION, Lipid-A-disaccharide synthase, SODIUM ION | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
5EFD
 
 | | Crystal structure of a surface pocket creating mutant (W6A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CHLORIDE ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2015-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.674 Å) | | Cite: | Small Glycols Discover Cryptic Pockets on Proteins for Fragment-Based Approaches.
J.Chem.Inf.Model., 2021
|
|
5EFF
 
 | | Crystal structure of an aromatic mutant (F4A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Beta-xylanase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2015-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of an aromatic mutant (F4A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27
To Be Published
|
|
5YBZ
 
 | |
6SJS
 
 | | Methyltransferase of the MtgA N227A mutant from Desulfitobacterium hafniense in complex with methyl-tetrahydrofolate | | Descriptor: | GLYCEROL, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, SODIUM ION, ... | | Authors: | Badmann, T, Groll, M. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures in Tetrahydrofolate Methylation in Desulfitobacterial Glycine Betaine Metabolism at Atomic Resolution.
Chembiochem, 21, 2020
|
|
