4ZQO
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor Q67 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
5HMK
 
 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {4-[2-(2-hydroxyethoxy)phenyl]piperazin-1-yl}[(2R,3S)-2-propyl-3-[4-(trifluoromethyl)phenoxy]-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}piperidin-3-yl]methanone | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
6TW4
 
 | | HumRadA22F in complex with compound 6 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6I3S
 
 | | Crystal structure of MDM2 in complex with compound 13. | | Descriptor: | (3~{S},3'~{R},3'~{a}~{S},6'~{a}~{R})-6-chloranyl-3'-(3-chloranyl-2-fluoranyl-phenyl)-1'-(cyclopropylmethyl)spiro[1~{H}-indole-3,2'-3~{a},6~{a}-dihydro-3~{H}-pyrrolo[3,4-b]pyrrole]-2,4'-dione, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | Authors: | Bader, G, Kessler, D. | | Deposit date: | 2018-11-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Targeted Synthesis of Complex Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-ones by Intramolecular Cyclization of Azomethine Ylides: Highly Potent MDM2-p53 Inhibitors.
ChemMedChem, 14, 2019
|
|
5HMM
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exodeoxyribonuclease, ... | | Authors: | Flemming, C.S, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6U1J
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ADP, phosphate, D-ala-D-ala, Mg2+ and K+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B, Thompson, A.P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
6U23
 
 | | EM structure of MPEG-1(w.t.) soluble pre-pore | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-19 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
6I55
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase (DHODH) co-crystallized with N-(2,2-Diphenylethyl)-4-hydroxy-1,2,5-thiadiazole-3-carboxamide | | Descriptor: | Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, N-(2,2-Diphenylethyl)-4-hydroxy-1,2,5-thiadiazole-3-carboxamide, ... | | Authors: | Goyal, P, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S. | | Deposit date: | 2018-11-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Hydroxyazole scaffold-based Plasmodium falciparum dihydroorotate dehydrogenase inhibitors: Synthesis, biological evaluation and X-ray structural studies.
Eur J Med Chem, 163, 2018
|
|
7BQK
 
 | | The structure of PdxI in complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 3-[(E,2S,4S)-2,4-dimethyloct-6-enoyl]-4-oxidanyl-1H-pyridin-2-one, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
6IDS
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN D35N mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
5HTZ
 
 | | BACE1 in complex with (S)-5-(3-chloro-5-(5-(prop-1-yn-1-yl)pyridin-3-yl)thiophen-2-yl)-2,5-dimethyl-1,2,4-thiadiazinan-3-iminium 1,1-dioxide | | Descriptor: | (3E,5S)-5-{3-chloro-5-[5-(prop-1-yn-1-yl)pyridin-3-yl]thiophen-2-yl}-2,5-dimethyl-1,2,4-thiadiazinan-3-imine 1,1-dioxide, Beta-secretase 1 | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
4Z7V
 
 | | L3-12 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DQ-alpha chain, MHC class II HLA-DQ-beta-1, ... | | Authors: | Petersen, J, Rossjohn, J, Reid, H.H, Koning, F. | | Deposit date: | 2015-04-08 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Determinants of Gliadin-Specific T Cell Selection in Celiac Disease.
J Immunol., 194, 2015
|
|
4Z8T
 
 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:AvrRxo1-ORF2 WITH SULPHATE IONS | | Descriptor: | ACETATE ION, AvrRxo1-ORF1, AvrRxo1-ORF2, ... | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z3O
 
 | | Quinolone(Moxifloxacin)-DNA cleavage complex of topoisomerase IV from S. pneumoniae | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA topoisomerase 4 subunit B,ParE30-ParC55 fused topo IV from S. pneumoniae, E-site DNA, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Selvarajah, J, Crevel, I.M.-T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2015-03-31 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
To Be Published
|
|
6TW9
 
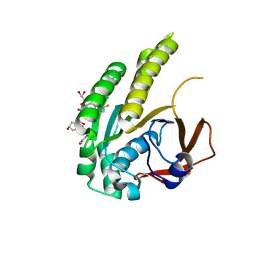 | | HumRadA22F in complex with CAM833 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
5HY2
 
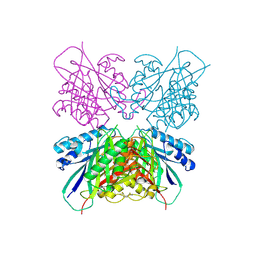 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
7BQL
 
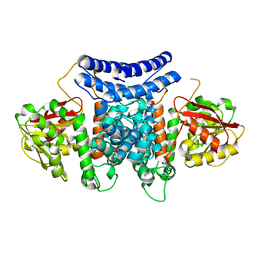 | | The crystal structure of PdxI complex with the Alder-ene adduct | | Descriptor: | 3-[(1R,2S,4R,6S)-2-ethenyl-4,6-dimethyl-cyclohexyl]-4-oxidanyl-1H-pyridin-2-one, DI(HYDROXYETHYL)ETHER, Methyltransf_2 domain-containing protein | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
4Z4J
 
 | |
5M27
 
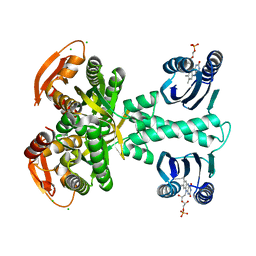 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-10-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
7BAV
 
 | | A de novo pentameric coiled-coil assembly: CC-Type2-(TgLaId)4-W19BrPhe. | | Descriptor: | CC-Type2-(TgLaId)4-W19BrPhe | | Authors: | Martin, F.J.O, Dawson, W.M, Shelley, K, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2020-12-16 | | Release date: | 2021-04-28 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Coiled coils 9-to-5: rational de novo design of alpha-helical barrels with tunable oligomeric states.
Chem Sci, 12, 2021
|
|
6U1E
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ATP, D-alanine-D-alanine, Mg2+ and Rb+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
5HX9
 
 | |
6U2L
 
 | | EM structure of MPEG-1 (L425K, beta conformation) soluble pre-pore complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
5LT1
 
 | |
6UBO
 
 | | Fluorogen Activating Protein Dib1 | | Descriptor: | 12-(diethylamino)-2,2-bis(fluoranyl)-4,5-dimethyl-5-aza-3-azonia-2-boranuidatricyclo[7.4.0.0^{3,7}]trideca-1(13),3,7,9,11-pentaen-6-one, CITRIC ACID, Outer membrane lipoprotein Blc, ... | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V.Z, Pletnev, S. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure-Based Rational Design of Two Enhanced Bacterial Lipocalin Blc Tags for Protein-PAINT Super-resolution Microscopy.
Acs Chem.Biol., 15, 2020
|
|
