1FSA
 
 | | THE T-STATE STRUCTURE OF LYS 42 TO ALA MUTANT OF THE PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Lu, G, Stec, B, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-08-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for an active T-state pig kidney fructose 1,6-bisphosphatase: interface residue Lys-42 is important for allosteric inhibition and AMP cooperativity.
Protein Sci., 5, 1996
|
|
7P6H
 
 | |
8ZUL
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8m | | Descriptor: | 2-azanyl-5-[2-[(3~{R})-3-azanylpyrrolidin-1-yl]pyridin-4-yl]-3-(2,6-dimethyl-3-oxidanyl-phenyl)benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.80026162 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZQA
 
 | |
8G5W
 
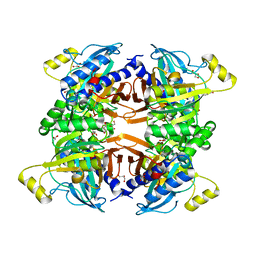 | | Structure of the Class II Fructose-1,6-Bisphophatase from Francisella tularensis complexed with native metal cofactor Mn++ | | Descriptor: | Fructose-1,6-bisphosphatase, GLYCEROL, MANGANESE (II) ION | | Authors: | Abad-Zapatero, C, Selezneva, A.I. | | Deposit date: | 2023-02-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New structures of Class II Fructose-1,6-Bisphosphatase from Francisella tularensis provide a framework for a novel catalytic mechanism for the entire class.
Plos One, 18, 2023
|
|
8GQ0
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with STL233497 | | Descriptor: | Bromodomain-containing protein 4, FORMIC ACID, GLYCEROL, ... | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2022-08-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
7PHH
 
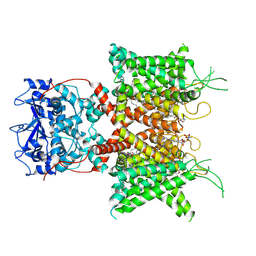 | | Human voltage-gated potassium channel Kv3.1 (apo condition) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Marsden, B, MacLean, E.M, Fernandez-Cid, A, Pike, A.C.W, Savva, C, Ragan, T.J, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
8GSS
 
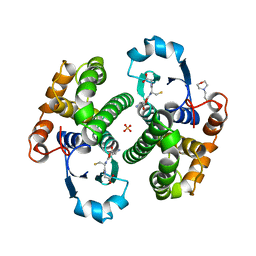 | | Human glutathione S-transferase P1-1, complex with glutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
8GN7
 
 | |
8Q5A
 
 | | Crystal structure of metal-dependent class II sulfofructosephosphate aldolase from Hafnia paralvei HpSqiA-Zn in complex with dihydroxyacetone phosphate (DHAP) | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Ketose-bisphosphate aldolase, ZINC ION | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
8KAP
 
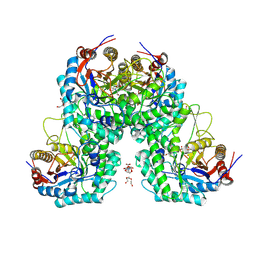 | | Glycoside hydrolase family 1 beta-glucosidase from Streptomyces griseus (ligand-free) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Kumakura, H, Motouchi, S, Nakai, H, Nakajima, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of beta-1,2-glucan-associated glycoside hydrolase family 1 beta-glucosidase from Streptomyces griseus
To Be Published
|
|
8EJ8
 
 | |
6UY8
 
 | |
8Z2W
 
 | | Crystal structure of apo- exo-beta-(1,3)-glucanase from Aspergillus oryzae (AoBgl) | | Descriptor: | 2-methylpropylphosphonic acid, Glucan 1,3-beta-glucosidase A, SODIUM ION | | Authors: | Banerjee, B, Kamale, C.K, Suryawanshi, A.B, Bhaumik, P. | | Deposit date: | 2024-04-13 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Aspergillus oryzae exo-beta-(1,3)-glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis.
Febs Lett., 599, 2025
|
|
6V3U
 
 | | Crystal Structure of the NDM_FIM-1 like Metallo-beta-Lactamase from Erythrobacter litoralis in the Mono-Zinc Form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase II, ISOPROPYL ALCOHOL, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-26 | | Release date: | 2020-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the NDM_FIM-1 like Metallo-beta-Lactamase from Erythrobacter litoralis in the Mono-Zinc Form
To Be Published
|
|
8F1C
 
 | | Voltage-gated potassium channel Kv3.1 with novel positive modulator (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one (compound 4) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one, POTASSIUM ION, ... | | Authors: | Chen, Y, Ishchenko, A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Identification and structural and biophysical characterization of a positive modulator of human Kv3.1 channels.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FA0
 
 | |
8YMH
 
 | | BRD4-BD1 in complex with 5-methyl-2-{[(2R)-2-methyl-4-methylsulfonyl-piperazin-1-yl]methyl}-7-(1-methylpyrazol-3-yl)furo[3,2-c]pyridin-4-one | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-2-{[(2R)-2-methyl-4-methylsulfonyl-piperazin-1-yl]methyl}-7-(1-methylpyrazol-3-yl)furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4 | | Authors: | Sasaki, C, Miyaguchi, I, Hagihara, S, Ishizawa, K, Endo, J. | | Deposit date: | 2024-03-09 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Discovery of a potent, orally available furopyridine derivative as a novel selective BET BD1 Inhibitor (Part 1).
To Be Published
|
|
1F5J
 
 | | CRYSTAL STRUCTURE OF XYNB, A HIGHLY THERMOSTABLE BETA-1,4-XYLANASE FROM DICTYOGLOMUS THERMOPHILUM RT46B.1, AT 1.8 A RESOLUTION | | Descriptor: | BETA-1,4-XYLANASE, SULFATE ION | | Authors: | McCarthy, A.A, Baker, E.N. | | Deposit date: | 2000-07-26 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of XynB, a highly thermostable beta-1,4-xylanase from Dictyoglomus thermophilum Rt46B.1, at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
6WBW
 
 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7PAV
 
 | | MALT1 in complex with compound 1 | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1, ~{N}1,~{N}4-bis[2-(trifluoromethyl)pyrimidin-4-yl]cyclohexane-1,4-diamine | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2021-07-30 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Discovery and optimization of cyclohexane-1,4-diamines as allosteric MALT1 inhibitors.
Eur.J.Med.Chem., 227, 2021
|
|
7PAW
 
 | | MALT1 in complex with compound 1 | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1, ~{N}1-(3-chloranyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-~{N}4-[2-(trifluoromethyl)pyrimidin-4-yl]cyclohexane-1,4-diamine | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2021-07-30 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery and optimization of cyclohexane-1,4-diamines as allosteric MALT1 inhibitors.
Eur.J.Med.Chem., 227, 2021
|
|
7OTX
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-257 | | Descriptor: | (S)-2-((3-(6-amino-9H-purin-9-yl)propyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OZ2
 
 | | Crystal structure of HIV-1 reverse transcriptase with a double stranded DNA showing a transient P-pocket | | Descriptor: | CADMIUM ION, DNA (28-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Das, K. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12, 2021
|
|
7OTN
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-247 | | Descriptor: | (S)-2-((2-(6-amino-9H-purin-9-yl)ethyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
