1J7E
 
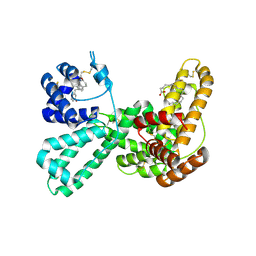 | | A Structural Basis for the Unique Binding Features of the Human Vitamin D-binding Protein | | Descriptor: | 3-(2-{4-[2-(5-HYDROXY-2-METHYLENE-CYCLOHEXYLIDENE)-ETHYLIDENE]-7A-METHYL-OCTAHYDRO-INDEN-1-YL}-PROPYL)-PHENOL, OLEIC ACID, vitamin D binding protein | | Authors: | Verboven, C, Rabijns, A, De Maeyer, M, Van Baelen, H, Bouillon, R, De Ranter, C. | | Deposit date: | 2001-05-16 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A structural basis for the unique binding features of the human vitamin D-binding protein.
Nat.Struct.Biol., 9, 2002
|
|
8ESC
 
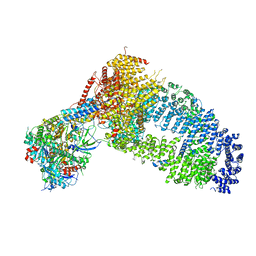 | | Structure of the Yeast NuA4 Histone Acetyltransferase Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Patel, A.B, Zukin, S.A, Nogales, E. | | Deposit date: | 2022-10-13 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and flexibility of the yeast NuA4 histone acetyltransferase complex.
Elife, 11, 2022
|
|
4K1N
 
 | |
1D1J
 
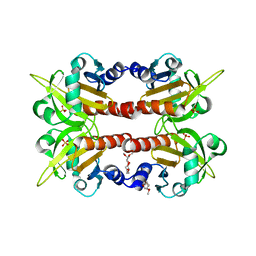 | | CRYSTAL STRUCTURE OF HUMAN PROFILIN II | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, PROFILIN II, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Lindberg, U, Schutt, C.E. | | Deposit date: | 1999-09-17 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure determination of human profilin II: A comparative structural analysis of human profilins.
J.Mol.Biol., 294, 1999
|
|
7ZI4
 
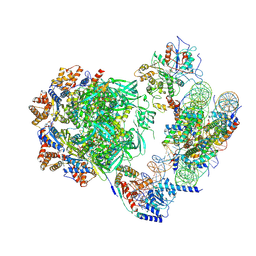 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
1CF0
 
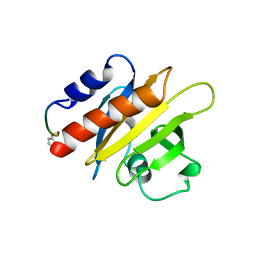 | |
5HVK
 
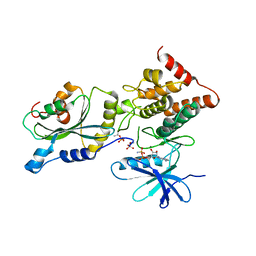 | |
4IGG
 
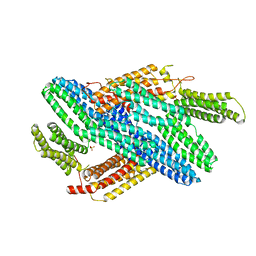 | |
6B0T
 
 | |
1T2L
 
 | | Three Crystal Structures of Human Coactosin-like Protein | | Descriptor: | Coactosin-like protein | | Authors: | Liu, L, Wei, Z, Chen, Z, Wang, Y, Gong, W. | | Deposit date: | 2004-04-22 | | Release date: | 2004-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Coactosin-like Protein
J.Mol.Biol., 344, 2004
|
|
1VFQ
 
 | |
8P5F
 
 | | Human wild-type GAPDH,orthorhombic form | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Samygina, V.R, Muronetz, V.I, Schmalhausen, E.V. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | S-nitrosylation and S-glutathionylation of GAPDH: Similarities, differences, and relationships.
Biochim Biophys Acta Gen Subj, 1867, 2023
|
|
8U95
 
 | | The structure of myosin heavy chain from Drosophila melanogaster flight muscle thick filaments | | Descriptor: | Myosin heavy chain, isoform U | | Authors: | Abbasi Yeganeh, F, Rastegarpouyani, H, Li, J, Taylor, K.A. | | Deposit date: | 2023-09-18 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the Drosophila melanogaster Flight Muscle Myosin Filament at 4.7 angstrom Resolution Reveals New Details of Non-Myosin Proteins.
Int J Mol Sci, 24, 2023
|
|
4YDH
 
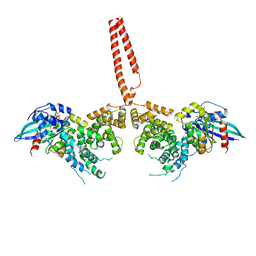 | | The structure of human FMNL1 N-terminal domains bound to Cdc42 | | Descriptor: | Cell division control protein 42 homolog, Formin-like protein 1, MAGNESIUM ION, ... | | Authors: | Kuhn, S, Anand, K, Geyer, M. | | Deposit date: | 2015-02-22 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation.
Nat Commun, 6, 2015
|
|
4TXI
 
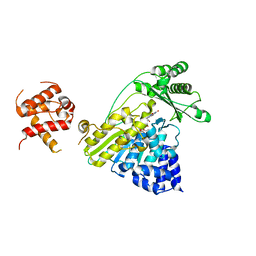 | | Construct of MICAL-1 containing the monooxygenase and calponin homology domains | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alqassim, S.S, Amzel, L.M, Bianchet, M.A. | | Deposit date: | 2014-07-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Modulation of MICAL Monooxygenase Activity by its Calponin Homology Domain: Structural and Mechanistic Insights.
Sci Rep, 6, 2016
|
|
6HTS
 
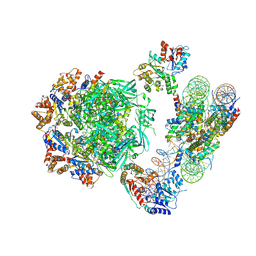 | | Cryo-EM structure of the human INO80 complex bound to nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Ayala, R, Willhoft, O, Aramayo, R.J, Wilkinson, M, McCormack, E.A, Ocloo, L, Wigley, D.B, Zhang, X. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and regulation of the human INO80-nucleosome complex.
Nature, 556, 2018
|
|
4YC7
 
 | |
8IYJ
 
 | | Cryo-EM structure of the 48-nm repeat doublet microtubule from mouse sperm | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhou, L.N, Gui, M, Wu, J.P. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of sperm flagellar doublet microtubules expand the genetic spectrum of male infertility.
Cell, 186, 2023
|
|
2W0P
 
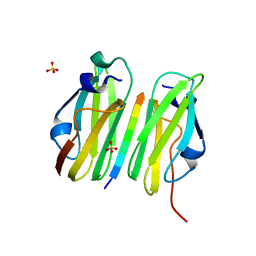 | |
8EUF
 
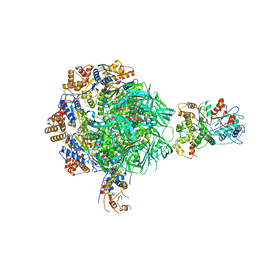 | | Class2 of the INO80-Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8W41
 
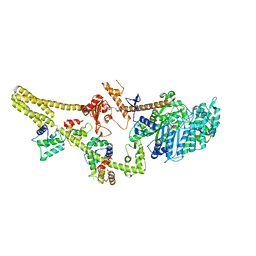 | | Cryo-EM structure of Myosin VI in the autoinhibited state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Calmodulin-1, ... | | Authors: | Niu, F, Wei, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Autoinhibition and activation of myosin VI revealed by its cryo-EM structure.
Nat Commun, 15, 2024
|
|
8EU9
 
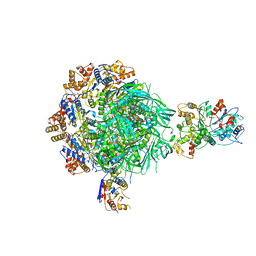 | | Class1 of the INO80-Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETU
 
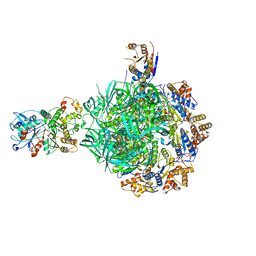 | | Class2 of the INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETS
 
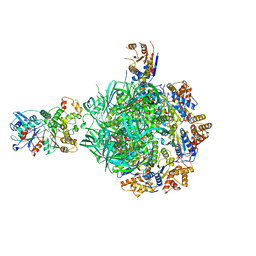 | | Class1 of the INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETW
 
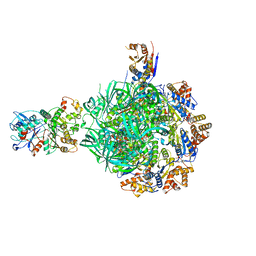 | | Class3 of INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
