2I05
 
 | | Escherichia Coli Replication Terminator Protein (Tus) Complexed With TerA DNA | | Descriptor: | 5'-D(*T*TP*AP*GP*TP*TP*AP*CP*AP*AP*CP*AP*TP*AP*CP*T)-3', 5'-D(*TP*AP*GP*TP*AP*TP*GP*TP*TP*GP*TP*AP*AP*CP*TP*A)-3', DNA replication terminus site-binding protein, ... | | Authors: | Oakley, A.J, Mulcair, M.D, Schaeffer, P.M, Dixon, N.E. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Polarity of Termination of DNA Replication in E. coli
To be Published
|
|
2I06
 
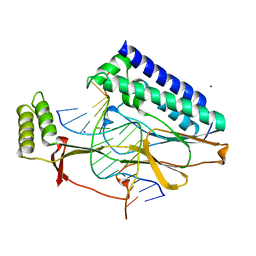 | | Escherichia Coli Replication Terminator Protein (Tus) Complexed With DNA- Locked form | | Descriptor: | 5'-D(*T*TP*AP*GP*TP*TP*AP*CP*AP*AP*CP*AP*TP*AP*CP*T)-3', 5'-D(*TP*G*AP*TP*AP*TP*GP*TP*TP*GP*TP*AP*AP*CP*TP*A)-3', DNA replication terminus site-binding protein, ... | | Authors: | Oakley, A.J, Mulcair, M.D, Schaeffer, P.M, Dixon, N.E. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Polarity of Termination of DNA Replication in E. coli.
To be Published
|
|
2I07
 
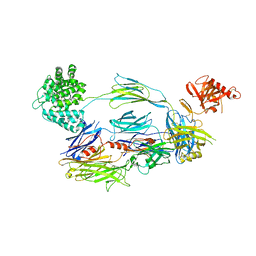 | | Human Complement Component C3b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3b, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Janssen, B.J.C, Christodoulidou, A, McCarthy, A, Lambris, J.D, Gros, P. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of C3b reveals conformational changes that underlie complement activity.
Nature, 444, 2006
|
|
2I08
 
 | |
2I0A
 
 | | Crystal Structure of KB-19 complexed with wild type HIV-1 protease | | Descriptor: | (5S)-3-(4-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | Deposit date: | 2006-08-10 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
2I0B
 
 | |
2I0C
 
 | |
2I0D
 
 | | Crystal structure of AD-81 complexed with wild type HIV-1 protease | | Descriptor: | (5S)-3-(3-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | Deposit date: | 2006-08-10 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
2I0E
 
 | |
2I0F
 
 | | Lumazine synthase RibH1 from Brucella abortus (Gene BruAb1_0785, Swiss-Prot entry Q57DY1) | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
2I0G
 
 | | Benzopyrans are Selective Estrogen Receptor beta Agonists (SERBAs) with Novel Activity in Models of Benign Prostatic Hyperplasia | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, Estrogen receptor beta | | Authors: | Wang, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Benzopyrans are selective estrogen receptor Beta agonists with novel activity in models of benign prostatic hyperplasia.
J.Med.Chem., 49, 2006
|
|
2I0H
 
 | | The structure of p38alpha in complex with an arylpyridazinone | | Descriptor: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2I0I
 
 | | X-ray crystal structure of Sap97 PDZ3 bound to the C-terminal peptide of HPV18 E6 | | Descriptor: | Disks large homolog 1, peptide E6 | | Authors: | Chen, X.S, Zhang, Y, Dasgupta, J, Banks, L, Thomas, M. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
J.Virol., 81, 2007
|
|
2I0J
 
 | | Benzopyrans are Selective Estrogen Receptor beta Agonists (SERBAs) with Novel Activity in Models of Benign Prostatic Hyperplasia | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, Estrogen receptor alpha | | Authors: | Wang, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Benzopyrans are selective estrogen receptor Beta agonists with novel activity in models of benign prostatic hyperplasia.
J.Med.Chem., 49, 2006
|
|
2I0K
 
 | |
2I0L
 
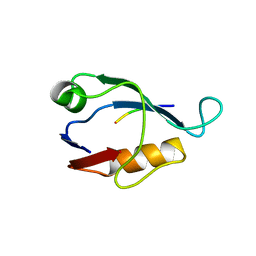 | | X-ray crystal structure of Sap97 PDZ2 bound to the C-terminal peptide of HPV18 E6. | | Descriptor: | Disks large homolog 1, peptide E6 | | Authors: | Chen, X.S, Zhang, Y, Dasgupta, J, Banks, L, Thomas, M. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
J.Virol., 81, 2007
|
|
2I0M
 
 | | Crystal structure of the phosphate transport system regulatory protein PhoU from Streptococcus pneumoniae | | Descriptor: | Phosphate transport system protein phoU, ZINC ION | | Authors: | Zhang, R, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-10 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the phosphate transport system regulatory protein PhoU from Streptococcus pneumoniae
To be Published, 2006
|
|
2I0N
 
 | | Structure of Dictyostelium discoideum Myosin VII SH3 domain with adjacent proline rich region | | Descriptor: | Class VII unconventional myosin | | Authors: | Wang, Q, Deloia, M.A, Kang, Y, Litchke, C, Titus, M.A, Walters, K.J. | | Deposit date: | 2006-08-10 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The SH3 domain of a M7 interacts with its C-terminal proline-rich region.
Protein Sci., 16, 2007
|
|
2I0O
 
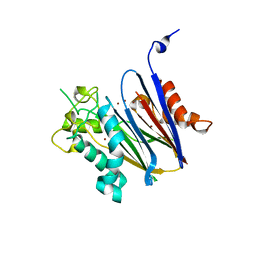 | | Crystal structure of Anopheles gambiae Ser/Thr phosphatase complexed with Zn2+ | | Descriptor: | Ser/Thr phosphatase, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2I0Q
 
 | |
2I0R
 
 | |
2I0S
 
 | |
2I0T
 
 | |
2I0U
 
 | |
2I0V
 
 | | c-FMS tyrosine kinase in complex with a quinolone inhibitor | | Descriptor: | 6-CHLORO-3-(3-METHYLISOXAZOL-5-YL)-4-PHENYLQUINOLIN-2(1H)-ONE, SULFATE ION, cFMS tyrosine kinase | | Authors: | Schubert, C, Schalk-Hihi, C. | | Deposit date: | 2006-08-11 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of colony-stimulating factor-1 receptor (cFMS) in complex with two inhibitors.
J.Biol.Chem., 282, 2007
|
|
