6NP8
 
 | |
4RZQ
 
 | | Structural Analysis of Substrate, Reaction Intermediate and Product Binding in Haemophilus influenzae Biotin Carboxylase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, methyl (3aS,4S,6aR)-4-(5-methoxy-5-oxopentyl)-2-oxohexahydro-1H-thieno[3,4-d]imidazole-1-carboxylate | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R, Waldrop, G.L. | | Deposit date: | 2014-12-23 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
6WYT
 
 | |
6NNP
 
 | |
6Z5S
 
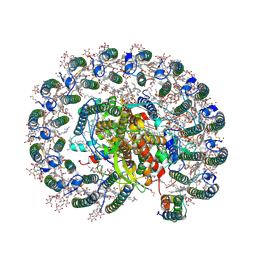 | | RC-LH1(14)-W complex from Rhodopseudomonas palustris | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, (6~{R},10~{S},14~{R},19~{R},23~{S},24~{E},27~{S},28~{E})-2,6,10,14,19,23,27,31-octamethyldotriaconta-24,28-dien-2-ol, ... | | Authors: | Swainsbury, D.J.K, Qian, P, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2020-05-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structures of Rhodopseudomonas palustris RC-LH1 complexes with open or closed quinone channels.
Sci Adv, 7, 2021
|
|
7Q13
 
 | | Human GYS1-GYG1 complex activated state bound to glucose-6-phosphate, uridine diphosphate, and glucose | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Glycogen [starch] synthase, muscle, ... | | Authors: | McCorvie, T.J, Shrestha, L, Froese, D.S, Ferreira, I.M, Yue, W.W. | | Deposit date: | 2021-10-17 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the regulation of human glycogen synthase by phosphorylation and glucose-6-phosphate.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8P2W
 
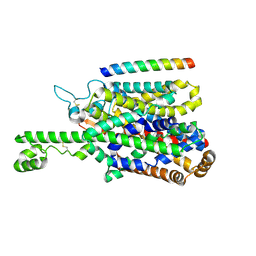 | | Structure of human SIT1 (focussed map / refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Sodium- and chloride-dependent transporter XTRP3 | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
7ZNR
 
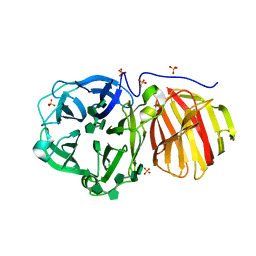 | |
6ZAV
 
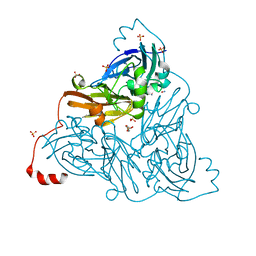 | | NO-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at 1.19 A resolution (unrestrained, full matrix refinement by SHELX) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
8CKO
 
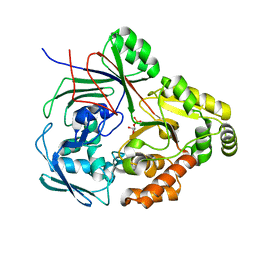 | | PBP AccA from A.tumefaciens C58 in complex with agrocinopine C-like | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ABC transporter substrate-binding protein, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8P2Z
 
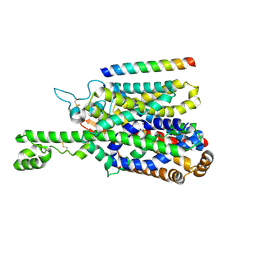 | | Structure of human SIT1 bound to L-pipecolate (focussed map / refinement) | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8CDO
 
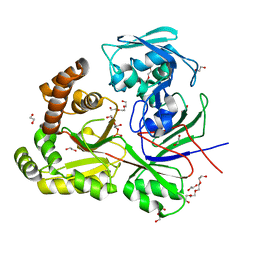 | |
7Q1M
 
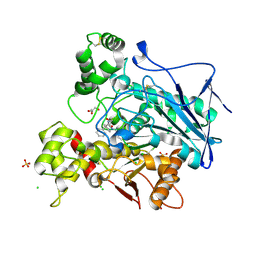 | | Crystal structure of human butyrylcholinesterase in complex with N-[(2S)-3-[(cyclohexylmethyl)amino]-2-hydroxypropyl]-2,2-diphenylacetamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Brazzolotto, X, Panek, D, Pasieka, A, Malawska, B, Nachon, F. | | Deposit date: | 2021-10-20 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of new, highly potent and selective inhibitors of BuChE - design, synthesis, in vitro and in vivo evaluation and crystallography studies.
Eur.J.Med.Chem., 249, 2023
|
|
8CBF
 
 | | SARS-CoV-2 Delta-RBD complexed with Omi-42 and Beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
7Q1N
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-[(2R)-3-[(cyclohexylmethyl)amino]-2-hydroxypropyl]-2,2-diphenylacetamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Brazzolotto, X, Panek, D, Pasieka, A, Malawska, B, Nachon, F. | | Deposit date: | 2021-10-20 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of new, highly potent and selective inhibitors of BuChE - design, synthesis, in vitro and in vivo evaluation and crystallography studies.
Eur.J.Med.Chem., 249, 2023
|
|
7Q1P
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-[(2R)-3-[(cyclohexylmethyl)amino]-2-hydroxypropyl]-3,3-diphenylpropanamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Brazzolotto, X, Panek, D, Pasieka, A, Malawska, B, Nachon, F. | | Deposit date: | 2021-10-20 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of new, highly potent and selective inhibitors of BuChE - design, synthesis, in vitro and in vivo evaluation and crystallography studies.
Eur.J.Med.Chem., 249, 2023
|
|
6NU1
 
 | | Crystal Structure of Human PKM2 in Complex with L-cysteine | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CYSTEINE, MAGNESIUM ION, ... | | Authors: | Srivastava, D, Nandi, S, Dey, M. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic and Structural Insights into Cysteine-Mediated Inhibition of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 58, 2019
|
|
6Z3J
 
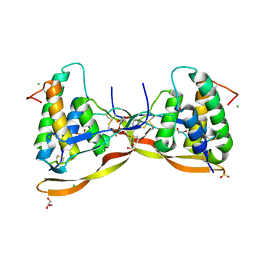 | | Repulsive Guidance Molecule B (RGMB) in complex with Growth Differentiation Factor 5 (GDF5) (crystal form 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Malinauskas, T, Peer, T.V, Bishop, B, Muller, T.D, Siebold, C. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Repulsive guidance molecules lock growth differentiation factor 5 in an inhibitory complex.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8ABP
 
 | |
4WMS
 
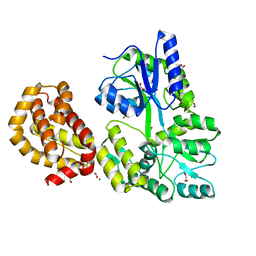 | | STRUCTURE OF APO MBP-MCL1 AT 1.9A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
7PBF
 
 | |
8FDP
 
 | |
8FLH
 
 | | Bruton's tyrosine kinase in complex with an orthosteric inhibitor | | Descriptor: | 2-(3,5-dichloroanilino)-1-{(3R)-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]azepan-1-yl}ethan-1-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of structural diverse reversible BTK inhibitors utilized to develop a novel in vivo CD69 and CD86 PK/PD mouse model.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
8FLG
 
 | | Bruton's tyrosine kinase in complex with an orthosteric inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, N~2~-(3-chlorophenyl)-N-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]glycinamide, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of structural diverse reversible BTK inhibitors utilized to develop a novel in vivo CD69 and CD86 PK/PD mouse model.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
6ZAU
 
 | | Damage-free nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
