6EKD
 
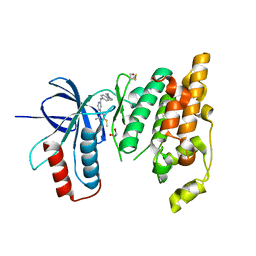 | | Crystal structure of JNK3 in complex with a pyridinylimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methyl-2-methylsulfanyl-1~{H}-imidazol-5-yl)-~{N}-(4-morpholin-4-ylphenyl)pyridin-2-amine, BETA-MERCAPTOETHANOL, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-09-26 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
1A2E
 
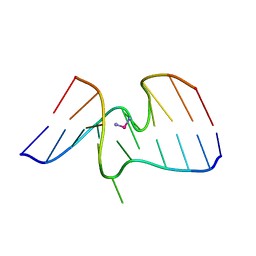 | |
3HD6
 
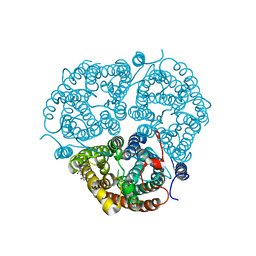 | | Crystal Structure of the Human Rhesus Glycoprotein RhCG | | Descriptor: | Ammonium transporter Rh type C, octyl beta-D-glucopyranoside | | Authors: | Gruswitz, F, Chaudhary, S, Ho, J.D, Pezeshki, B, Ho, C.-M, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-05-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Function of human Rh based on structure of RhCG at 2.1 A.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5Z9Z
 
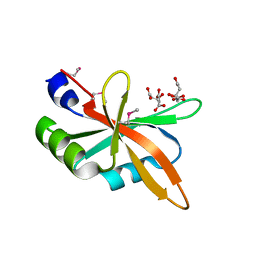 | | The C-terminal RRM domain of Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 (E329A/E330A/E332A) | | Descriptor: | CITRATE ANION, Small RNA degrading nuclease 1 | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Cao, C, Gan, J, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
2B55
 
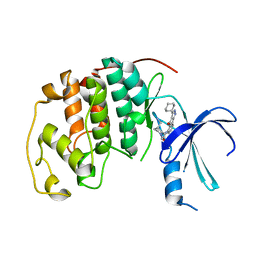 | | Human cyclin dependent kinase 2 (cdk2) complexed with indenopyraxole DIN-101312 | | Descriptor: | 2-(4-(AMINOMETHYL)PIPERIDIN-1-YL)-N-(3_CYCLOHEXYL-4-OXO-2,4-DIHYDROINDENO[1,2-C]PYRAZOL-5-YL)ACETAMIDE, Cell division protein kinase 2 | | Authors: | Muckelbauer, J. | | Deposit date: | 2005-09-27 | | Release date: | 2005-10-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and Evaluation of Indenopyrazoles as Cyclin-Dependent Kinase Inhibitors. 3. Structure Activity Relationships at C3
J.Med.Chem., 45, 2002
|
|
7DMW
 
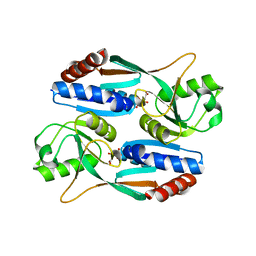 | | Crystal structure of CcpC regulatory domain in complex with citrate from Bacillus amyloliquefaciens | | Descriptor: | CITRATE ANION, CcpC | | Authors: | Chen, J, Wang, L, Shang, F, Liu, W, Chen, Y, Lan, J, Bu, T, Bai, X, Xu, Y. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and structural analysis of catabolite control protein C that responds to citrate.
Sci Rep, 11, 2021
|
|
3GAX
 
 | | Crystal structure of monomeric human cystatin C stabilized against aggregation | | Descriptor: | Cystatin-C | | Authors: | Kolodziejczyk, R, Michalska, K, Hernandez-Santoyo, A, Wahlbom, M, Grubb, A, Jaskolski, M. | | Deposit date: | 2009-02-18 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human cystatin C stabilized against amyloid formation.
Febs J., 277, 2010
|
|
2IZ8
 
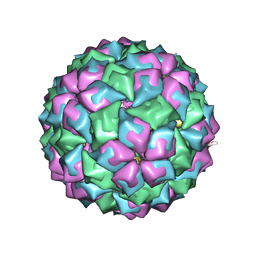 | | MS2-RNA HAIRPIN (C-7) COMPLEX | | Descriptor: | MS2 COAT PROTEIN, RNA, (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*CP*UP*CP* AP*CP*CP*CP*AP*UP*GP*U)-3') | | Authors: | Helgstrand, C, Grahn, E, Stonehouse, N.J, Stockley, P.G, Liljas, L. | | Deposit date: | 2006-07-25 | | Release date: | 2006-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Investigating the Structural Basis of Purine Specificity in the Structures of MS2 Coat Protein RNA Translational Operator Hairpins
Nucleic Acids Res., 30, 2002
|
|
5EBM
 
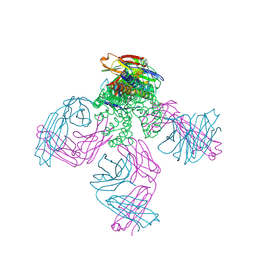 | | KcsA T75G mutant in the nonconductive state | | Descriptor: | Antibody Fab Fragment Light Chain, DIACYL GLYCEROL, NONAN-1-OL, ... | | Authors: | Matulef, K, Valiyaveetil, F.I. | | Deposit date: | 2015-10-19 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Individual Ion Binding Sites in the K(+) Channel Play Distinct Roles in C-type Inactivation and in Recovery from Inactivation.
Structure, 24, 2016
|
|
4BDB
 
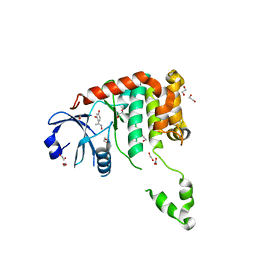 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]benzene-1,3-diol, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
6KU7
 
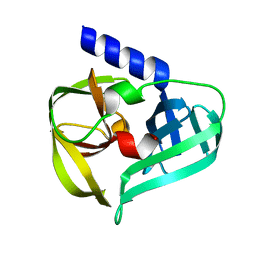 | | structure of HRV-C 3C protein | | Descriptor: | Genome polyprotein | | Authors: | Zhu, L, Yuan, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the HRV-C 3C-Rupintrivir Complex Provides New Insights for Inhibitor Design.
Virol Sin, 35, 2020
|
|
2X6M
 
 | | Structure of a single domain camelid antibody fragment in complex with a C-terminal peptide of alpha-synuclein | | Descriptor: | ALPHA-SYNUCLEIN PEPTIDE, HEAVY CHAIN VARIABLE DOMAIN FROM DROMEDARY | | Authors: | DeGenst, E, Guilliams, T, Wellens, J, O'Day, E.M, Waudby, C.A, Meehan, S, Dumoulin, M, Hsu, S.-T.D, Cremades, N, Verschueren, K.H.G, Pardon, E, Wyns, L, Steyaert, J, Christodoulou, J, Dobson, C.M. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Properties of a Complex of Alpha-Synuclein and a Single-Domain Camelid Antibody.
J.Mol.Biol., 402, 2010
|
|
5I9H
 
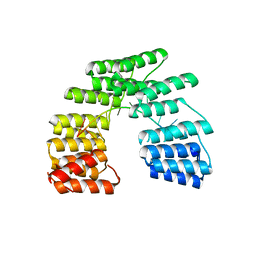 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8G2 in complex with its target RNA U8G2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*GP*GP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8G2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
1RRS
 
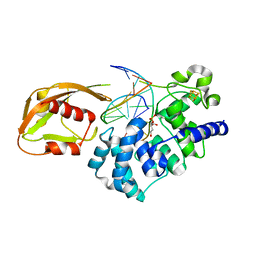 | | MutY adenine glycosylase in complex with DNA containing an abasic site | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
5L3L
 
 | | D11 bound IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
5L3M
 
 | | D11 bound [S39_PQ]-IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
5L3N
 
 | | D11 bound [N29, S39_PQ]-IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
6EZI
 
 | |
5I9G
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8C2 in complex with its target RNA U8C2 | | Descriptor: | RNA (5'-R(*GP*GP*G*GP*UP*UP*UP*UP*CP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8C2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5ICF
 
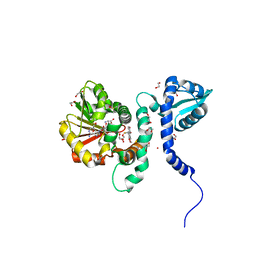 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine and sanguinarine | | Descriptor: | (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
3HL5
 
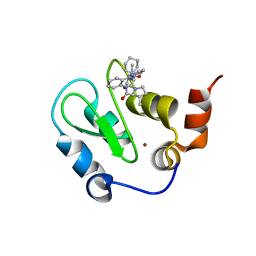 | | Crystal structure of XIAP BIR3 with CS3 | | Descriptor: | (3S)-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-3-methyl-N-(2-pyrimidin-2-ylphenyl)-L-prolinamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Hymowitz, S.G. | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism of c-IAP and XIAP proteins is required for efficient induction of cell death by small-molecule IAP antagonists.
Acs Chem.Biol., 4, 2009
|
|
7EAO
 
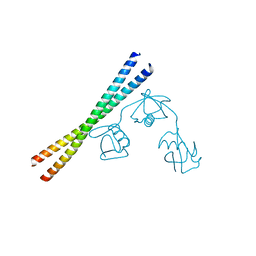 | |
3HRO
 
 | | Crystal structure of a C-terminal coiled coil domain of Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2, polycystic kidney disease 2) | | Descriptor: | Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2), also called Polycystin-2 or polycystic kidney disease 2(PKD2) | | Authors: | Yu, Y, Ulbrich, M.H, Li, M.-H, Buraei, Z, Chen, X.-Z, Ong, A.C.M, Tong, L, Isacoff, E.Y, Yang, J. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and molecular basis of the assembly of the TRPP2/PKD1 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2JEG
 
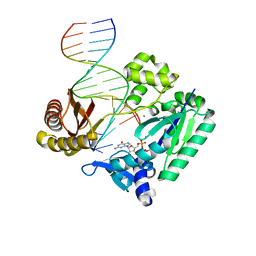 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*DOC)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JEI
 
 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*T)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
