5EM4
 
 | | Structure of CYP2B4 F244W in a ligand free conformation | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shah, M.B, Stout, C.D, Halpert, J.R. | | Deposit date: | 2015-11-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Coumarin Derivatives as Substrate Probes of Mammalian Cytochromes P450 2B4 and 2B6: Assessing the Importance of 7-Alkoxy Chain Length, Halogen Substitution, and Non-Active Site Mutations.
Biochemistry, 55, 2016
|
|
6MVV
 
 | | NavAb voltage-gated sodium channel, I217C/F203A | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein, PHOSPHATE ION | | Authors: | Lenaeus, M.J, Catterall, W.A. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fenestrations control resting-state block of a voltage-gated sodium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6MUH
 
 | | Fluoroacetate dehalogenase, room temperature structure solved by serial 1 degree oscillation crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
6N01
 
 | | Structure of apo AztD from Citrobacter koseri | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AztD Protein, ... | | Authors: | Yukl, E.T, Neupane, D.P. | | Deposit date: | 2018-11-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of AztD provide mechanistic insights into direct zinc transfer between proteins.
Commun Biol, 2, 2019
|
|
3X2N
 
 | | Proton relay pathway in inverting cellulase | | Descriptor: | Endoglucanase V-like protein, SULFATE ION | | Authors: | Nakamura, A, Ishida, T, Fushinobu, S, Igarashi, K, Samejima, M. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
4DUU
 
 | |
1S5Z
 
 | | NDP kinase in complex with adenosine phosphonoacetic acid | | Descriptor: | ADENOSINE PHOSPHONOACETIC ACID, Nucleoside diphosphate kinase, cytosolic, ... | | Authors: | Chen, Y, Morera, S, Pasti, C, Angusti, A, Solaroli, N, Veron, M, Janin, J, Manfredini, S, Deville-Bonne, D. | | Deposit date: | 2004-01-22 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Adenosine phosphonoacetic acid is slowly metabolized by NDP kinase.
Med.Chem., 1, 2005
|
|
2LK5
 
 | | Solution structure of the Zn(II) form of Desulforedoxin | | Descriptor: | Desulforedoxin, ZINC ION | | Authors: | Goodfellow, B.J, Tavares, P, Romao, M.J, Czaja, C, Rusnak, F, Legall, J, Moura, I, Moura, J.J.G. | | Deposit date: | 2011-10-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of desulforedoxin, a simple iron-sulfur protein - An NMR study of the zinc derivative
J.BIOL.INORG.CHEM., 1, 1996
|
|
1GD3
 
 | | refined solution structure of human cystatin A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix
conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
4YMK
 
 | | Crystal Structure of Stearoyl-Coenzyme A Desaturase 1 | | Descriptor: | Acyl-CoA desaturase 1, STEAROYL-COENZYME A, ZINC ION, ... | | Authors: | Bai, Y, McCoy, J.G, Rajashankar, K.R, Zhou, M. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | X-ray structure of a mammalian stearoyl-CoA desaturase.
Nature, 524, 2015
|
|
9FK9
 
 | | The structure of XT6 from G.proteiniphilus T-6: The E265G/Q238A/W241A mutant | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, ZINC ION, ... | | Authors: | Hadad, N, Chmelnik, O, Dessau, M, Shoham, Y, Shoham, G. | | Deposit date: | 2024-06-02 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of XT6 from G.proteiniphilus T-6: The E265G/Q238A/W241A mutant
To Be Published
|
|
6FW2
 
 | | Crystal Structure of human mARC1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MOLYBDATE ION, Mitochondrial amidoxime-reducing component 1,Endolysin,Mitochondrial amidoxime-reducing component 1, ... | | Authors: | Kubitza, C, Scheidig, A. | | Deposit date: | 2018-03-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human mARC1 reveals its exceptional position among eukaryotic molybdenum enzymes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8HTU
 
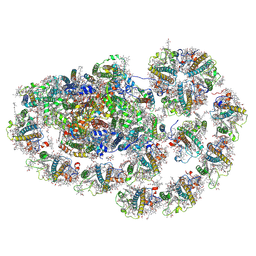 | | Cryo-EM structure of PpPSI-L | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Li, M, Pan, X.W, Sun, H.Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into the assembly and energy transfer of the Lhcb9-dependent photosystem I from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
6DUI
 
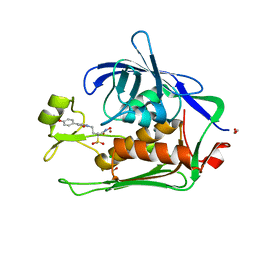 | |
6DGT
 
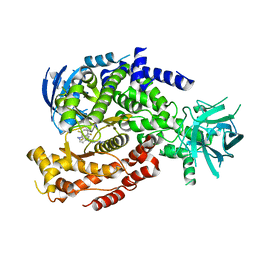 | | Selective PI3K beta inhibitor bound to PI3K delta | | Descriptor: | 4-[1-(5,8-difluoroquinolin-4-yl)-2-methyl-4-(4H-1,2,4-triazol-3-yl)-1H-benzimidazol-6-yl]-3-fluoropyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J, Villasenor, A, McGrath, M. | | Deposit date: | 2018-05-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Atropisomerism by Design: Discovery of a Selective and Stable Phosphoinositide 3-Kinase (PI3K) beta Inhibitor.
J. Med. Chem., 61, 2018
|
|
9CJQ
 
 | | X-ray crystal structure of SARS-CoV-2 main protease quadruple mutants in complex with Ensitrelvir | | Descriptor: | 1,2-ETHANEDIOL, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, main protease | | Authors: | Esler, M.A, Shi, K, Harris, R.S, Aihara, H. | | Deposit date: | 2024-07-07 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | X-ray crystal structure of SARS-CoV-2 main protease quadruple mutants in complex with Ensitrelvir
To Be Published
|
|
4Q73
 
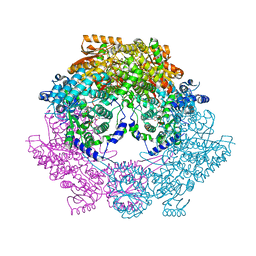 | | Crystal Structure of Bradyrhizobium japonicum Proline Utilization A (PutA) Mutant D778Y | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Proline dehydrogenase, ... | | Authors: | Tanner, J.J, Luo, M, Pemberton, T.A. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A.
Biochemistry, 53, 2014
|
|
9CJU
 
 | | Structure of SARS-CoV-2 main protease in complex with Bofutrelvir in orthorhombic form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Non-structural protein 7, ... | | Authors: | Esler, M.A, Shi, K, Harris, R.S, Aihara, H. | | Deposit date: | 2024-07-07 | | Release date: | 2025-07-09 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for varying drug resistance of SARS-CoV-2 M pro E166 variants.
Mbio, 16, 2025
|
|
6GK9
 
 | | Inhibited structure of IMPDH from Pseudomonas aeruginosa | | Descriptor: | (5~{S})-7-azanyl-5-(4-chlorophenyl)-2,4-bis(oxidanylidene)-1,5-dihydropyrano[2,3-d]pyrimidine-6-carbonitrile, Inosine-5'-monophosphate dehydrogenase, SULFATE ION | | Authors: | Labesse, G, Alexandre, T, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2018-05-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | First-in-class allosteric inhibitors of bacterial IMPDHs.
Eur J Med Chem, 167, 2019
|
|
6T9R
 
 | | Aplysia californica AChBP in complex with a cytisine derivative | | Descriptor: | (1~{R},9~{S})-5-(3-oxidanylpropyl)-7,11-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4-dien-6-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine binding protein, ... | | Authors: | Davis, S, Hunter, W.N. | | Deposit date: | 2019-10-28 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The thermodynamic profile and molecular interactions of a C(9)-cytisine derivative-binding acetylcholine-binding protein from Aplysia californica.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6D7K
 
 | | Complex structure of Methane monooxygenase hydroxylase in complex with inhibitory subunit | | Descriptor: | FE (III) ION, FORMIC ACID, HEXANE-1,6-DIOL, ... | | Authors: | Kim, H, Lee, S.J, Cho, U.-S. | | Deposit date: | 2018-04-24 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MMOD-induced structural changes of hydroxylase in soluble methane monooxygenase.
Sci Adv, 5, 2019
|
|
6D9R
 
 | | The substrate-bound crystal structure of HPRT (hypoxanthine phosphoribosyltransferase) | | Descriptor: | 1,2-ETHANEDIOL, 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAGUANINE, ... | | Authors: | Satyshur, K.A, Wolak, C, Anderson, B, Dubiel, K, Keck, J.L. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Evolution of (p)ppGpp-HPRT regulation through diversification of an allosteric oligomeric interaction.
Elife, 8, 2019
|
|
8CMO
 
 | | Cryo-EM structure of the Photosystem I - LHCI supercomplex from Coelastrella sp. | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Fadeeva, M, Klaiman, D, Nelson, N. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of the Photosystem I - LHCI supercomplex from Coelastrella sp.
To Be Published
|
|
6DBC
 
 | | Second bromodomain of Human BRD2 with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2S,4R)-4-[(2-chlorophenyl)amino]-2-methyl-6-(1H-pyrazol-3-yl)-3,4-dihydroquinolin-1(2H)-yl]ethan-1-one, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-03 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Second bromodomain of Human BRD2 with a Tetrahydroquinoline analogue
To Be Published
|
|
4CCS
 
 | | The structure of CbiX, the terminal Enzyme for Biosynthesis of Siroheme in Denitrifying Bacteria | | Descriptor: | 1,2-ETHANEDIOL, CBIX, D-MALATE, ... | | Authors: | Bali, S, Rollauer, S.E, Roversi, P, Raux-Deery, E, Lea, S.M, Warren, M.J, Ferguson, S.J. | | Deposit date: | 2013-10-25 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of the 'Missing' Terminal Enzyme for Siroheme Biosynthesis in Alpha-Proteobacteria.
Mol.Microbiol., 92, 2014
|
|
