5MXY
 
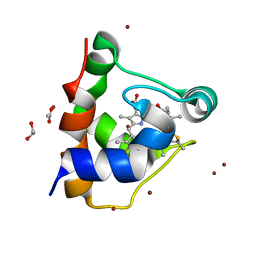 | | KustC0563 c-type cytochrome | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cytochrome c-552 Ks_3358, ... | | Authors: | Mohd, A, Barends, T. | | Deposit date: | 2017-01-25 | | Release date: | 2018-02-14 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Specificity of Small c -Type Cytochromes in Anaerobic Ammonium Oxidation.
Acs Omega, 6, 2021
|
|
5MY4
 
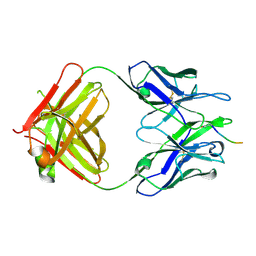 | |
5VN9
 
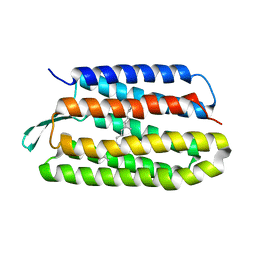 | | Structure of bacteriorhodopsin from crystals grown at 4 deg C using GlyNCOC15+4 as an LCP host lipid | | Descriptor: | Bacteriorhodopsin | | Authors: | Ishchenko, A, Peng, L, Zinovev, E, Vlasov, A, Lee, S.C, Kuklin, A, Mishin, A, Borshchevskiy, V, Zhang, Q, Cherezov, V. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Chemically Stable Lipids for Membrane Protein Crystallization.
Cryst Growth Des, 17, 2017
|
|
7LGR
 
 | |
6DF4
 
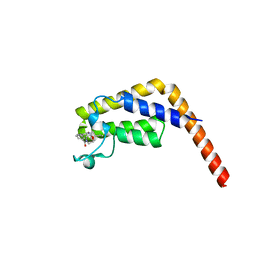 | | TAF1-BD2 in complex with Cpd8 (6-(but-3-en-1-yl)-4-(3-(morpholine-4-carbonyl)phenyl)-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one) | | Descriptor: | 6-(but-3-en-1-yl)-4-[3-(morpholine-4-carbonyl)phenyl]-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Transcription initiation factor TFIID subunit | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2018-05-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | GNE-371, a Potent and Selective Chemical Probe for the Second Bromodomains of Human Transcription-Initiation-Factor TFIID Subunit 1 and Transcription-Initiation-Factor TFIID Subunit 1-like.
J. Med. Chem., 61, 2018
|
|
6DF7
 
 | |
5K64
 
 | | Crystal structure of VEGF binding IgG1-Fc (Fcab 448) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Humm, A, Lobner, E, Kitzmuller, M, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
8SM9
 
 | |
3LRR
 
 | | Crystal structure of human RIG-I CTD bound to a 12 bp AU rich 5' ppp dsRNA | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*(ATP)P*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*U)-3'), ZINC ION | | Authors: | Li, P. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structural Basis of 5' Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain.
Structure, 18, 2010
|
|
5VX6
 
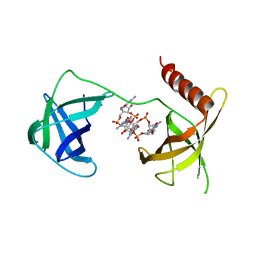 | | Structure of Bacillus subtilis Inhibitor of motility (MotI/DgrA) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Uncharacterized protein YpfA | | Authors: | Subramanian, S, Dann III, C. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | MotI (DgrA) acts as a molecular clutch on the flagellar stator protein MotA inBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6DJR
 
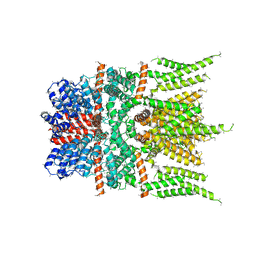 | |
5YTF
 
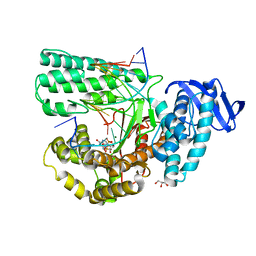 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
1N31
 
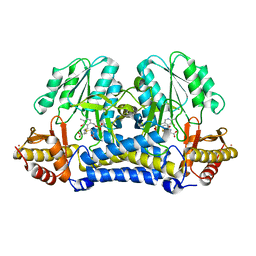 | | Structure of A Catalytically Inactive Mutant (K223A) of C-DES with a Substrate (Cystine) Linked to the Co-Factor | | Descriptor: | CYSTEINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
7JWE
 
 | |
8TB5
 
 | | TYK2 JH2 bound to Compound7 | | Descriptor: | ACETATE ION, N-{(3P)-3-[3-(dimethylsulfamoyl)phenyl]-1H-pyrrolo[2,3-c]pyridin-5-yl}cyclopropanecarboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Argiriadi, M.A, Van Epps, S.A, Breinlinger, E.C. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Targeting the Tyrosine Kinase 2 (TYK2) Pseudokinase Domain: Discovery of the Selective TYK2 Inhibitor ABBV-712.
J.Med.Chem., 66, 2023
|
|
8TB6
 
 | | TYK2 JH2 bound to Compound14 | | Descriptor: | N-[(3M)-3-{6-[(3R)-3-methoxyoxolan-3-yl]pyridin-2-yl}-1-methyl-1H-pyrrolo[2,3-c]pyridin-5-yl]urea, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Argiriadi, M.A, Van Epps, S.A, Breinlinger, E.C. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting the Tyrosine Kinase 2 (TYK2) Pseudokinase Domain: Discovery of the Selective TYK2 Inhibitor ABBV-712.
J.Med.Chem., 66, 2023
|
|
4JQD
 
 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OL operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*CP*AP*CP*TP*AP*AP*GP*GP*AP*AP*AP*AP*CP*TP*TP*AP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*AP*AP*GP*TP*TP*TP*TP*CP*CP*TP*TP*AP*GP*TP*GP*T)-3') | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4OT9
 
 | | crystal structure of the C-terminal domain of p100/NF-kB2 | | Descriptor: | Nuclear factor NF-kappa-B p100 subunit, SULFATE ION | | Authors: | Tao, Z.H, Huang, D.B, Fusco, A, Gupta, K, Ware, C.F, Duynne, G.V. | | Deposit date: | 2014-02-13 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | p100/I kappa B delta sequesters and inhibits NF-kappa B through kappaBsome formation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1L9B
 
 | | X-Ray Structure of the Cytochrome-c(2)-Photosynthetic Reaction Center Electron Transfer Complex from Rhodobacter sphaeroides in Type II Co-Crystals | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CHLORIDE ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Okamura, M.Y, Yeh, A.P, Rees, D.C, Feher, G. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure determination of the cytochrome c2: reaction center electron transfer complex from Rhodobacter sphaeroides.
J.Mol.Biol., 319, 2002
|
|
1L2G
 
 | | Structure of a C-terminally truncated form of glycoprotein D from HSV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein D | | Authors: | Carfi, A, Willis, S.H, Whitbeck, J.C, Krummenacher, C, Cohen, G.H, Eisenberg, R.J, Wiley, D.C. | | Deposit date: | 2002-02-21 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Herpes simplex virus glycoprotein D bound to the human receptor HveA.
Mol.Cell, 8, 2001
|
|
7Z2I
 
 | | TRYPSIN (BOVINE) COMPLEXED WITH compound 4 | | Descriptor: | 5-[[3-(trifluoromethyl)phenyl]methyl]-1,4,6,7-tetrahydroimidazo[4,5-c]pyridine, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schiering, N, Dalvit, C, Vulpetti, A. | | Deposit date: | 2022-02-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Efficient Screening of Target-Specific Selected Compounds in Mixtures by 19 F NMR Binding Assay with Predicted 19 F NMR Chemical Shifts.
Chemmedchem, 17, 2022
|
|
5OUJ
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 39 | | Descriptor: | 2-[(1~{R})-5-(4-chlorophenyl)-9-fluoranyl-3-methyl-1-oxidanyl-1~{H}-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
6KU8
 
 | | structure of HRV-C 3C protein with rupintrivir | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, Genome polyprotein | | Authors: | Zhu, L, Yuan, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the HRV-C 3C-Rupintrivir Complex Provides New Insights for Inhibitor Design.
Virol Sin, 35, 2020
|
|
5DFR
 
 | |
2WY6
 
 | | Clostridium perfringens alpha-toxin strain NCTC8237 mutant T74I | | Descriptor: | CADMIUM ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Vachieri, S.G, Naylor, C.E, Basak, A.K. | | Deposit date: | 2009-11-12 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Comparison of a Nontoxic Variant of Clostridium Perfringens [Alpha]-Toxin with the Toxic Wild-Type Strain
Acta Crystallogr.,Sect.D, 66, 2011
|
|
