2BS6
 
 | | LECTIN FROM RALSTONIA SOLANACEARUM COMPLEXED WITH XYLOGLUCAN FRAGMENT | | Descriptor: | GLYCEROL, LECTIN, alpha-L-fucopyranose, ... | | Authors: | Mitchell, E.P, Kostlanova, N, Wimmerova, M, Imberty, A. | | Deposit date: | 2005-05-18 | | Release date: | 2005-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The fucose-binding lectin from Ralstonia solanacearum. A new type of beta-propeller architecture formed by oligomerization and interacting with fucoside, fucosyllactose, and plant xyloglucan.
J. Biol. Chem., 280, 2005
|
|
7EK0
 
 | | Complex Structure of antibody BD-503 and RBD-N501Y of COVID-19 | | Descriptor: | Heavy Chain of BD-503, Light Chain of BD-503, Spike protein S1 | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJZ
 
 | | Complex Structure of antibody BD-503 and RBD-S477N of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJY
 
 | | Complex Structure of antibody BD-503 and RBD of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
3J8Z
 
 | |
3F52
 
 | | Crystal structure of the clp gene regulator ClgR from C. glutamicum | | Descriptor: | GLYCEROL, clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
2BWR
 
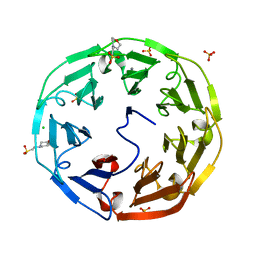 | | Crystal Structure of Psathyrella Velutina Lectin at 1.5A Resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | Deposit date: | 2005-07-18 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
2C25
 
 | | 1.8A Crystal Structure of Psathyrella velutina lectin in complex with N-acetylneuraminic acid | | Descriptor: | CALCIUM ION, N-acetyl-alpha-neuraminic acid, PSATHYRELLA VELUTINA LECTIN PVL, ... | | Authors: | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | Deposit date: | 2005-09-26 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
2BS5
 
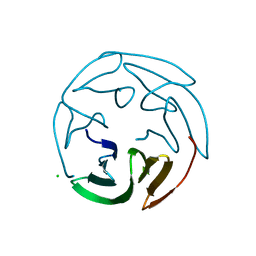 | | LECTIN FROM RALSTONIA SOLANACEARUM COMPLEXED WITH 2-FUCOSYLLACTOSE | | Descriptor: | CHLORIDE ION, FUCOSE-BINDING LECTIN PROTEIN, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Mitchell, E.P, Kostlanova, N, Wimmerova, M, Imberty, A. | | Deposit date: | 2005-05-18 | | Release date: | 2005-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Fucose-Binding Lectin from Ralstonia Solanacearum: A New Type of {Beta}-Propeller Architecture Formed by Oligomerization and Interacting with Fucoside, Fucosyllactose, and Plant Xyloglucan.
J.Biol.Chem., 280, 2005
|
|
2BWM
 
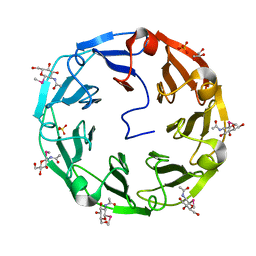 | | 1.8A CRYSTAL STRUCTURE OF of Psathyrella velutina LECTIN IN COMPLEX WITH METHYL 2-ACETAMIDO-1,2-DIDEOXY-1-SELENO-BETA-D-GLUCOPYRANOSIDE | | Descriptor: | CALCIUM ION, GLYCEROL, PSATHYRELLA VELUTINA LECTIN PVL, ... | | Authors: | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | Deposit date: | 2005-07-15 | | Release date: | 2006-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
2C4D
 
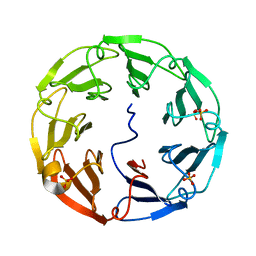 | | 2.6A Crystal Structure of Psathyrella velutina Lectin in Complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PSATHYRELLA VELUTINA LECTIN PVL, ... | | Authors: | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | Deposit date: | 2005-10-18 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
3J8W
 
 | |
3J8V
 
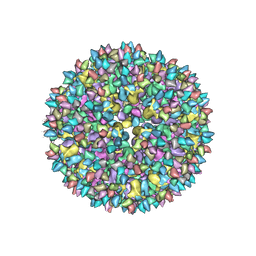 | |
4BXU
 
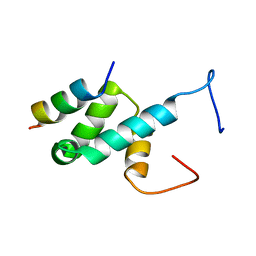 | | Structure of Pex14 in complex with Pex5 LVxEF motif | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Kooshapur, H, Meyer, H.N, Madl, T, Sattler, M. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Novel Pex14 Interacting Site of Human Pex5 is Critical for Matrix Protein Import Into Peroxisomes.
J.Biol.Chem., 289, 2014
|
|
7BB5
 
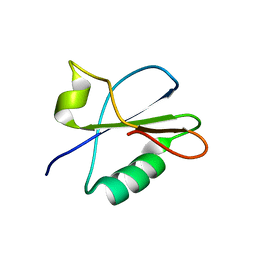 | |
3F51
 
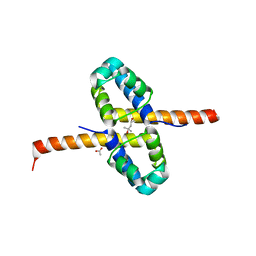 | | Crystal Structure of the clp gene regulator ClgR from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
6FXU
 
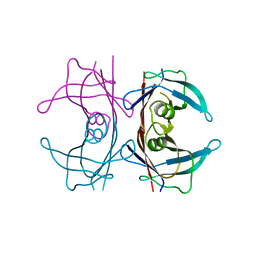 | |
6FZL
 
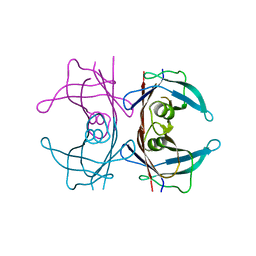 | |
7WN2
 
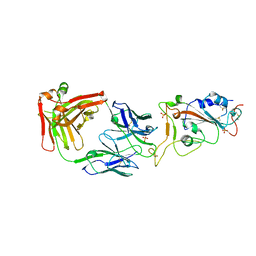 | |
7WNB
 
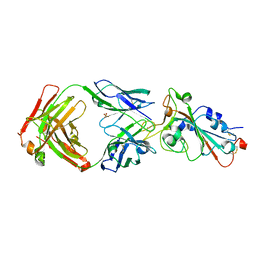 | |
7X87
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with sophotetraose observed as sophorose | | Descriptor: | Beta-galactosidase, CALCIUM ION, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J Biol Chem, 298, 2022
|
|
3TGX
 
 | | IL-21:IL21R complex | | Descriptor: | Interleukin-21, Interleukin-21 receptor, NICKEL (II) ION, ... | | Authors: | Hamming, O.J, Kang, L, Svenson, A, Karlsen, J.L, Rahbek-Nielsen, H, Paludan, S.R, Hjort, S.A, Bondensgaard, K, Hartmann, R. | | Deposit date: | 2011-08-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the interleukin 21 receptor bound to interleukin 21 reveals that a sugar chain interacting with the WSXWS motif is an integral part of the interleukin 21 receptor.
J.Biol.Chem., 2012
|
|
6FWD
 
 | |
7F6Z
 
 | | Complex Structure of antibody BD-503 and RBD-501Y.V2 of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7F6Y
 
 | | Complex Structure of antibody BD-503 and RBD-E484K of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
