4WPZ
 
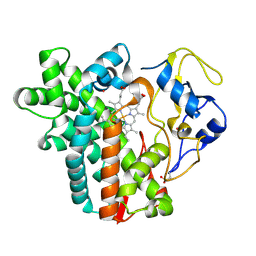 | | Crystal structure of cytochrome P450 CYP107W1 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kang, L.W, Kim, D.H, Pham, T.V, Han, S.H. | | Deposit date: | 2014-10-21 | | Release date: | 2015-04-22 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional characterization of CYP107W1 from Streptomyces avermitilis and biosynthesis of macrolide oligomycin A.
Arch.Biochem.Biophys., 575, 2015
|
|
1MK1
 
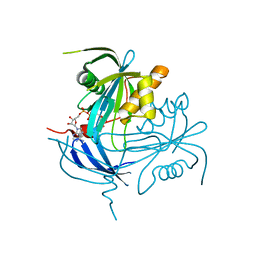 | | Structure of the MT-ADPRase in complex with ADPR, a Nudix enzyme | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADPR pyrophosphatase | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-08-28 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1MP2
 
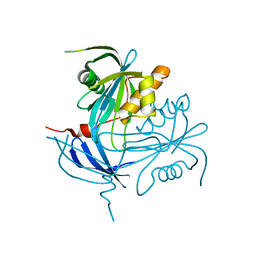 | | Structure of MT-ADPRase (Apoenzyme), a Nudix hydrolase from Mycobacterium tuberculosis | | Descriptor: | ADPR pyrophosphatase | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1MQE
 
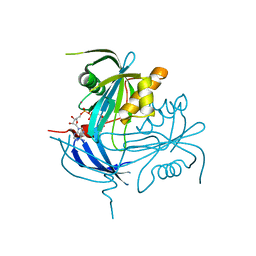 | | Structure of the MT-ADPRase in complex with gadolidium and ADP-ribose, a Nudix enzyme | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADPR pyrophosphatase, GADOLINIUM ION | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1MR2
 
 | | Structure of the MT-ADPRase in complex with 1 Mn2+ ion and AMP-CP (a inhibitor), a nudix enzyme | | Descriptor: | ADPR pyrophosphatase, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1MQW
 
 | | Structure of the MT-ADPRase in complex with three Mn2+ ions and AMPCPR, a Nudix enzyme | | Descriptor: | ADPR pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, MANGANESE (II) ION | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1NQZ
 
 | | The structure of a CoA pyrophosphatase from D. Radiodurans complexed with a magnesium ion | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein), MAGNESIUM ION | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
1NQY
 
 | | The structure of a CoA pyrophosphatase from D. Radiodurans | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein) | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
2O4N
 
 | | Crystal Structure of HIV-1 Protease (TRM Mutant) in Complex with Tipranavir | | Descriptor: | GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, protease | | Authors: | Kang, L.W, Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
2O4P
 
 | | Crystal Structure of HIV-1 Protease (Q7K) in Complex with Tipranavir | | Descriptor: | GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, protease | | Authors: | Kang, L.W, Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
4WQ0
 
 | |
3HE8
 
 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B | | Descriptor: | GLYCEROL, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
3HEE
 
 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B and ribose-5-phosphate | | Descriptor: | RIBOSE-5-PHOSPHATE, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
3E6G
 
 | | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae | | Descriptor: | Cystathionine gamma-lyase-like protein | | Authors: | Ngo, H.P.T, Kim, J.K, Kim, H.S, Jung, J.H, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, H.W, Kang, L.W. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae
To be published
|
|
3P14
 
 | |
3UU0
 
 | | Crystal structure of L-rhamnose isomerase from Bacillus halodurans in complex with Mn | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-27 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UVA
 
 | | Crystal structure of L-rhamnose isomerase mutant W38F from Bacillus halodurans in complex with Mn | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Jeya, M, Kim, J.K, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UXI
 
 | | Crystal structure of L-rhamnose isomerase W38A mutant from Bacillus halodurans | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
1MSN
 
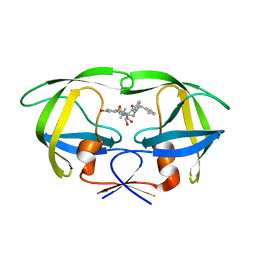 | | The HIV protease (mutant Q7K L33I L63I V82F I84V) complexed with KNI-764 (an inhibitor) | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
5I8U
 
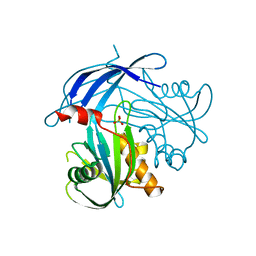 | | Crystal Structure of the RV1700 (MT ADPRASE) E142Q mutant | | Descriptor: | ADP-ribose pyrophosphatase, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Thirawatananond, P, Kang, L.-W, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and mutational studies of the adenosine diphosphate ribose hydrolase from Mycobacterium tuberculosis.
J. Bioenerg. Biomembr., 48, 2016
|
|
4Y21
 
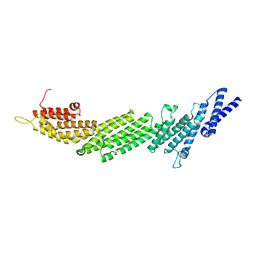 | | Crystal Structure of Munc13-1 MUN domain | | Descriptor: | Protein unc-13 homolog A | | Authors: | Yang, X.Y, Wang, S, Sheng, Y, Zhang, M, Zou, W.J, Wu, L.J, Kang, L.J, Rizo, J, Zhang, R.G, Xu, T, Ma, C. | | Deposit date: | 2015-02-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3TGX
 
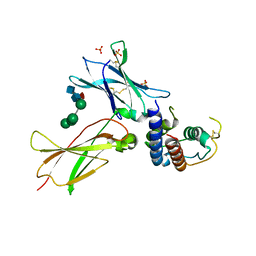 | | IL-21:IL21R complex | | Descriptor: | Interleukin-21, Interleukin-21 receptor, NICKEL (II) ION, ... | | Authors: | Hamming, O.J, Kang, L, Svenson, A, Karlsen, J.L, Rahbek-Nielsen, H, Paludan, S.R, Hjort, S.A, Bondensgaard, K, Hartmann, R. | | Deposit date: | 2011-08-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the interleukin 21 receptor bound to interleukin 21 reveals that a sugar chain interacting with the WSXWS motif is an integral part of the interleukin 21 receptor.
J.Biol.Chem., 2012
|
|
3DKU
 
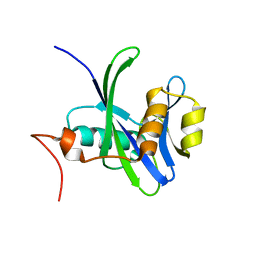 | | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1 | | Descriptor: | Putative phosphohydrolase | | Authors: | Hong, M.K, Kim, J.K, Jung, J.H, Jung, J.W, Choi, J.Y, Kang, L.W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1.
To be Published
|
|
1MRX
 
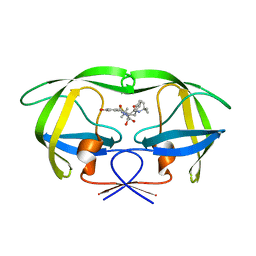 | | Structure of HIV protease (Mutant Q7K L33I L63I V82F I84V ) complexed with KNI-577 | | Descriptor: | (4R)-N-tert-butyl-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-1,3-thiazoli dine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
1MRW
 
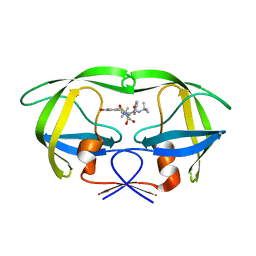 | | Structure of HIV protease (Mutant Q7K L33I L63I) complexed with KNI-577 | | Descriptor: | (4R)-N-tert-butyl-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-1,3-thiazoli dine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
