2R45
 
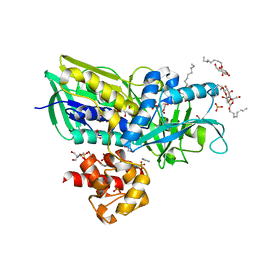 | | Crystal structure of Escherichia coli Glycerol-3-phosphate Dehydrogenase in complex with 2-phospho-d-glyceric acid | | Descriptor: | 1,2-ETHANEDIOL, 2-PHOSPHOGLYCERIC ACID, Aerobic glycerol-3-phosphate dehydrogenase, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential
monotopic membrane enzyme involved in respiration and metabolism
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CJT
 
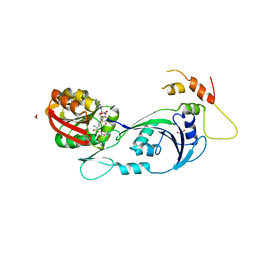 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with dimethylated ribosomal protein L11 | | Descriptor: | 1,2-ETHANEDIOL, 50S ribosomal protein L11, CHLORIDE ION, ... | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
3FL5
 
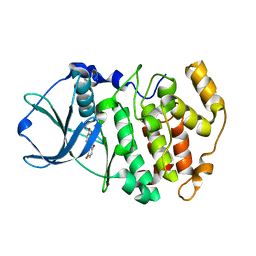 | | Protein kinase CK2 in complex with the inhibitor Quinalizarin | | Descriptor: | 1,2,5,8-tetrahydroxyanthracene-9,10-dione, Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER | | Authors: | Mazzorana, M, Franchin, C, Battistutta, R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quinalizarin as a potent, selective and cell-permeable inhibitor of protein kinase CK2
Biochem.J., 421, 2009
|
|
2QO1
 
 | | 2.6 Angstrom Crystal Structure of the Complex Between 11-(decyldithiocarbonyloxy)-undecanoic acid and Mycobacterium Tuberculosis FabH. | | Descriptor: | 11-[(MERCAPTOCARBONYL)OXY]UNDECANOIC ACID, 3-oxoacyl-[acyl-carrier-protein] synthase 3, DECANE-1-THIOL | | Authors: | Sachdeva, S, Musayev, F, Alhamadsheh, M, Scarsdale, J.N, Wright, H.T, Reynolds, K.A. | | Deposit date: | 2007-07-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Separate Entrance and Exit Portals for Ligand Traffic in Mycobacterium tuberculosis FabH
Chem.Biol., 15, 2008
|
|
2QQG
 
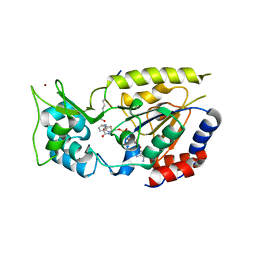 | | Hst2 bound to ADP-HPD, acetyllated histone H4 and nicotinamide | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Histone H4, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B, Zhao, K, Slama, J. | | Deposit date: | 2007-07-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
3FXQ
 
 | | Crystal structure of the LysR-type transcriptional regulator TsaR | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
3FYI
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides in the reduced state bound with cyanide | | Descriptor: | CADMIUM ION, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Qin, L, Mills, D.A, Proshlyakov, D.A, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redox dependent conformational changes in cytochrome c oxidase suggest a gating mechanism for proton uptake.
Biochemistry, 48, 2009
|
|
3GGS
 
 | | Human purine nucleoside phosphorylase double mutant E201Q,N243D complexed with 2-fluoro-2'-deoxyadenosine | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Sawaya, M.R, Afshar, S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of a mutant human purine nucleoside phosphorylase with the prodrug, 2-fluoro-2'-deoxyadenosine and the cytotoxic drug, 2-fluoroadenine.
Protein Sci., 18, 2009
|
|
3CMU
 
 | |
3BB1
 
 | | Crystal structure of Toc34 from Pisum sativum in complex with Mg2+ and GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Koenig, P, Sinning, I, Schleiff, E, Tews, I. | | Deposit date: | 2007-11-09 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GTPase cycle of the chloroplast import receptors Toc33/Toc34: implications from monomeric and dimeric structures.
Structure, 16, 2008
|
|
2RBA
 
 | | Structure of Human Thymine DNA Glycosylase Bound to Abasic and Undamaged DNA | | Descriptor: | DNA (5'-D(*DCP*DAP*DGP*DCP*DTP*DCP*DTP*DGP*DTP*DAP*DCP*DGP*DTP*DGP*DAP*DGP*DCP*DAP*DGP*DTP*DGP*DGP*DA)-3'), DNA (5'-D(*DCP*DCP*DAP*DCP*DTP*DGP*DCP*DTP*DCP*DAP*(3DR)P*DGP*DTP*DAP*DCP*DAP*DGP*DAP*DGP*DCP*DTP*DGP*DT)-3'), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Maiti, A, Pozharski, E, Drohat, A.C. | | Deposit date: | 2007-09-18 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2R7O
 
 | | Crystal Structure of VP1 apoenzyme of Rotavirus SA11 (N-terminal hexahistidine-tagged) | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-09 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
3G7F
 
 | | Crystal structure of Blastochloris viridis heterodimer mutant reaction center | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Ponomarenko, N.S, Li, L, Tereshko, V, Ismagilov, R.F, Norris Jr, J.R. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and spectropotentiometric analysis of Blastochloris viridis heterodimer mutant reaction center
Biochim.Biophys.Acta, 1788, 2009
|
|
2R9S
 
 | | c-Jun N-terminal Kinase 3 with 3,5-Disubstituted Quinoline inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 10, N-(tert-butyl)-4-[5-(pyridin-2-ylamino)quinolin-3-yl]benzenesulfonamide, ... | | Authors: | Habel, J. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3,5-Disubstituted quinolines as novel c-Jun N-terminal kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3BDM
 
 | | yeast 20S proteasome:glidobactin A-complex | | Descriptor: | (2E,4E)-N-[(2S,3R)-3-hydroxy-1-[[(3Z,5S,8S,10S)-10-hydroxy-5-methyl-2,7-dioxo-1,6-diazacyclododec-3-en-8-yl]amino]-1-ox obutan-2-yl]dodeca-2,4-dienamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Dudler, R, Kaiser, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism
Nature, 452, 2008
|
|
3BBX
 
 | |
3DFY
 
 | | Crystal structure of apo dipeptide epimerase from Thermotoga maritima | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-12 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening.
Structure, 16, 2008
|
|
2RDZ
 
 | | High Resolution Crystal Structure of the Escherichia coli Cytochrome c Nitrite Reductase. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Hemmings, A.M, RIchardson, D.J. | | Deposit date: | 2007-09-25 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
3FSO
 
 | |
3G0W
 
 | |
3G6K
 
 | | Crystal Structure of Candida glabrata FMN Adenylyltransferase in complex with FAD and Inorganic Pyrophosphate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FMN adenylyltransferase, MAGNESIUM ION, ... | | Authors: | Huerta, C, Machius, M, Zhang, H. | | Deposit date: | 2009-02-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and mechanism of a eukaryotic FMN adenylyltransferase.
J.Mol.Biol., 389, 2009
|
|
3FVR
 
 | |
3FYT
 
 | |
2RCE
 
 | |
2REK
 
 | | Crystal structure of tetR-family transcriptional regulator | | Descriptor: | ACETATE ION, Putative tetR-family transcriptional regulator | | Authors: | Dong, A, Xu, X, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-26 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of tetR-family transcriptional regulator.
To be Published
|
|
