7Z3B
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, reduced form | | Descriptor: | ACETATE ION, AcoP, COPPER (I) ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7AVB
 
 | |
7Z3F
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, oxidized form | | Descriptor: | ACETATE ION, AcoP, CHLORIDE ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3G
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, H166A mutant | | Descriptor: | AcoP, COPPER (I) ION, GLYCEROL | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3I
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, M171A mutant | | Descriptor: | ACETATE ION, AcoP, COPPER (II) ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
4K7Z
 
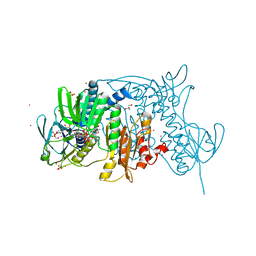 | | Crystal structure of the C136(42)A/C141(47)A double mutant of Tn501 MerA in complex with NADP and Hg2+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Dong, A, Falkowaski, M, Malone, M, Miller, S.M, Pai, E.F. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the C136(42)A/C141(47)A double mutant of Tn501 MerA in complex with NADP and Hg2+
To be Published
|
|
7B1E
 
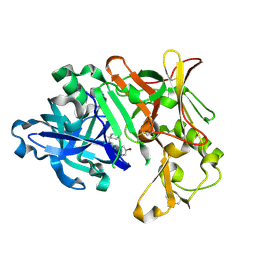 | | BACE1 IN COMPLEX WITH compound 3 (NB-641) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(4~{S})-2-azanyl-4-methyl-5,6-dihydro-1,3-thiazin-4-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7B1Q
 
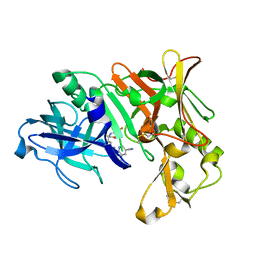 | | Crystal Structure of Human BACE-1 in Complex with Compound NB-360 (compound 54) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]-4-fluoranyl-phenyl]-5-cyano-3-methyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-25 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7T8Y
 
 | | Structure of Class A sortase from Streptococcus pyogenes bound to lipid II mimetic, LPATA: Thr-in conformation | | Descriptor: | ACE-ALA-ZGL-LYS-DAL-DAL, BE2-LEU-PRO-ALA-THR-ALA-ALA, Sortase | | Authors: | Piper, I.M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-12-17 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition.
J.Biol.Chem., 298, 2022
|
|
7T8Z
 
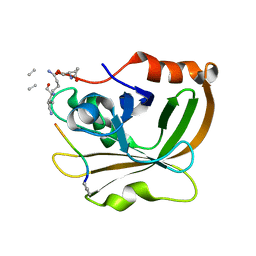 | | Structure of Class A sortase from Streptococcus pyogenes bound to lipid II mimetic, LPATA: Thr-out conformation | | Descriptor: | ACE-ALA-ZGL-LYS-DAL-DAL, BE2-LEU-PRO-ALA-THR-ALA-ALA, Sortase | | Authors: | Piper, I.M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-12-17 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition.
J.Biol.Chem., 298, 2022
|
|
7AV6
 
 | | FAST in a domain-swapped dimer form | | Descriptor: | FORMIC ACID, Photoactive yellow protein | | Authors: | Bukhdruker, S, Remeeva, A, Ruchkin, D, Gorbachev, D, Povarova, N, Mineev, K, Goncharuk, S, Baranov, M, Mishin, A, Borshchevskiy, V. | | Deposit date: | 2020-11-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NanoFAST: structure-based design of a small fluorogen-activating protein with only 98 amino acids.
Chem Sci, 12, 2021
|
|
6YMQ
 
 | | TREM2 extracellular domain (19-131) in complex with single-chain variable 4 (scFv-4) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Single-chain variable 4, ... | | Authors: | Szykowska, A, Preger, C, Scacioc, A, Mukhopadhyay, S.M.M, McKinley, G, Graslund, S, Wigren, E, Persson, H, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Di Daniel, E, Davis, J.B, Burgess-Brown, N, Bullock, A. | | Deposit date: | 2020-04-09 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
3VKS
 
 | | Assimilatory nitrite reductase (Nii3) - NO complex from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRIC OXIDE, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
7ANK
 
 | | Crystal structure of sarcomeric protein FATZ-1 (d91-FATZ-1 construct) in complex with half dimer of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-10-12 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
6Y6C
 
 | | TREM2 extracellular domain (19-174) in complex with single-chain variable fragment (scFv-4) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single chain variable, Triggering receptor expressed on myeloid cells 2 | | Authors: | Szykowska, A, Preger, C, Williams, E, Mukhopadhyay, S.M.M, McKinley, G, Gruslund, S, Wigren, E, Persson, H, Arrowsmith, C.H, Edwards, A, von Delft, F, Bountra, C, Davis, J.B, Di Daniel, E, Burgess-Brown, N, Bullock, A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
5OWK
 
 | |
7AT5
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a1 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, Casein kinase II subunit alpha, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
3IJS
 
 | | Structure of S67-27 in Complex with TSBP | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-alpha-D-glucopyranose, Immunoglobulin heavy chain (IGG3), Immunoglobulin light chain (IGG3), ... | | Authors: | Brooks, C.L, Blackler, R.J, Evans, S.V. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen.
Glycobiology, 20, 2010
|
|
7ATV
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha', ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-31 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
4K8D
 
 | | Crystal structure of the C558(464)A/C559(465)A double mutant of Tn501 MerA in complex with NADPH and Hg2+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase, ... | | Authors: | Dong, A, Falkowski, M, Malone, M, Miller, S.M, Pai, E.F. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the C136(42)A/C141(47)A double mutant of Tn501 MerA in complex with NADPH and
Hg2+
to be published
|
|
7AWI
 
 | | Crystal structure of human butyrylcholinesterase in complex with tert-butyl 3-(((2-((1-benzyl-1H-indol-4-yl)oxy)ethyl)amino)methyl]piperidine-1-carboxylate | | Descriptor: | 2-((1-benzyl-1H-indol-4-yl)oxy)-N-((1-(tert-butoxycarbonyl)piperidin-3-yl)methyl)ethan-1-aminium, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Goral, I, Wieckowska, A. | | Deposit date: | 2020-11-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development and crystallography-aided SAR studies of multifunctional BuChE inhibitors and 5-HT 6 R antagonists with beta-amyloid anti-aggregation properties.
Eur.J.Med.Chem., 225, 2021
|
|
7AWH
 
 | | Crystal structure of human butyrylcholinesterase in complex with tert-butyl 3-(((2-((1-(benzenesulfonyl)-1H-indol-4-yl)oxy)ethyl)amino)methyl)piperidine-1-carboxylate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Wichur, T, Wieckowska, A. | | Deposit date: | 2020-11-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development and crystallography-aided SAR studies of multifunctional BuChE inhibitors and 5-HT 6 R antagonists with beta-amyloid anti-aggregation properties.
Eur.J.Med.Chem., 225, 2021
|
|
7AHR
 
 | | Enzyme of biosynthetic pathway | | Descriptor: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase, GLYCEROL | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
8A18
 
 | | 1.63 A resolution hydroquinone inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Ciurli, S, Cianci, M. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Inhibition of Urease by Hydroquinones: A Structural and Kinetic Study.
Chemistry, 28, 2022
|
|
2IG2
 
 | | DIR PRIMAERSTRUKTUR DES KRISTALLISIERBAREN MONOKLONALEN IMMUNOGLOBULINS IGG1 KOL. II. AMINOSAEURESEQUENZ DER L-KETTE, LAMBDA-TYP, SUBGRUPPE I (GERMAN) | | Descriptor: | IGG1-LAMBDA KOL FAB (HEAVY CHAIN), IGG1-LAMBDA KOL FAB (LIGHT CHAIN) | | Authors: | Marquart, M, Huber, R. | | Deposit date: | 1989-04-18 | | Release date: | 1989-07-12 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The primary structure of crystallizable monoclonal immunoglobulin IgG1 Kol. II. Amino acid sequence of the L-chain, gamma-type, subgroup I
Biol.Chem.Hoppe-Seyler, 370, 1989
|
|
