3UE3
 
 | | Crystal structure of Acinetobacter baumanni PBP3 | | Descriptor: | Septum formation, penicillin binding protein 3, peptidoglycan synthetase | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3OUV
 
 | |
6DR3
 
 | | Crystal structure of E. coli LpoA amino terminal domain | | Descriptor: | Penicillin-binding protein activator LpoA | | Authors: | Kelley, A.C, Saper, M.A. | | Deposit date: | 2018-06-11 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structures of the amino-terminal domain of LpoA from Escherichia coli and Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7LQ6
 
 | | CryoEM structure of Escherichia coli PBP1b | | Descriptor: | Penicillin-binding protein 1B | | Authors: | Caveney, N.A, Workman, S.D, Yan, R, Atkinson, C.E, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | CryoEM structure of the antibacterial target PBP1b at 3.3 angstrom resolution.
Nat Commun, 12, 2021
|
|
3NB6
 
 | |
3NB7
 
 | |
2MHK
 
 | | E. coli LpoA N-terminal domain | | Descriptor: | Penicillin-binding protein activator LpoA | | Authors: | Jean, N.L, Bougault, C, Lodge, A, Derouaux, A, Callens, G, Egan, A, Lewis, R.J, Vollmer, W, Simorre, J. | | Deposit date: | 2013-11-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Elongated Structure of the Outer-Membrane Activator of Peptidoglycan Synthesis LpoA: Implications for PBP1A Stimulation.
Structure, 22, 2014
|
|
2IWB
 
 | | MecR1 unbound extracellular antibiotic-sensor domain. | | Descriptor: | GLYCEROL, METHICILLIN RESISTANCE MECR1 PROTEIN, NICKEL (II) ION, ... | | Authors: | Marrero, A, Mallorqui-Fernandez, G, Guevara, T, Garcia-Castellanos, R, Gomis-Ruth, F.X. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unbound and Acylated Structures of the Mecr1 Extracellular Antibiotic-Sensor Domain Provide Insights Into the Signal-Transduction System that Triggers Methicillin Resistance.
J.Mol.Biol., 361, 2006
|
|
2IWD
 
 | | Oxacilloyl-acylated MecR1 extracellular antibiotic-sensor domain. | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Methicillin resistance mecR1 protein | | Authors: | Marrero, A, Mallorqui-Fernandez, G, Guevara, T, Garcia-Castellanos, R, Gomis-Ruth, F.X. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unbound and Acylated Structures of the Mecr1 Extracellular Antibiotic-Sensor Domain Provide Insights Into the Signal-Transduction System that Triggers Methicillin Resistance.
J.Mol.Biol., 361, 2006
|
|
7ONN
 
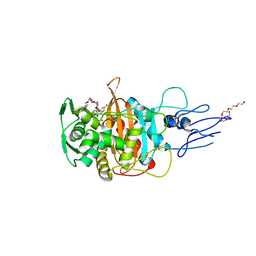 | | Crystal structure of PBP3 transpeptidase domain from E. coli in complex with AIC499 | | Descriptor: | (2S)-2-[(Z)-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2S)-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxy-3-[4-[N-[(3R)-piperidin-3-yl]carbamimidoyl]phenoxy]propanoic acid, DODECAETHYLENE GLYCOL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
4WEJ
 
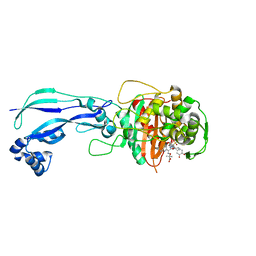 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted allyl monocarbam | | Descriptor: | (3R,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-[(5R)-5-hydroxy-4-oxo-4,5-dihydropyridin-2-yl]-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-3-(prop-2-en-1-yl)-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4WEL
 
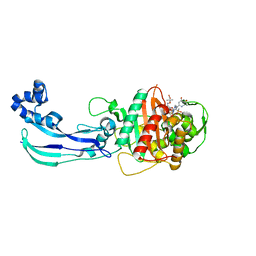 | | Crystal structure of Pseudomonas aeruginosa PBP3 with SMC-3176 | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-3,10,10-trimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4WEK
 
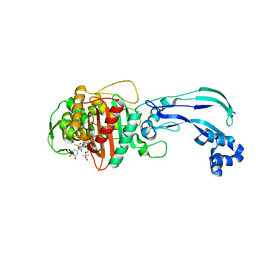 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted vinyl monocarbam | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-3-ethenyl-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
7ONO
 
 | |
7ONW
 
 | | Crystal structure of PBP3 from E. coli in complex with AIC499 | | Descriptor: | (2S)-2-[(Z)-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2S)-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxy-3-[4-[N-[(3R)-piperidin-3-yl]carbamimidoyl]phenoxy]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONY
 
 | | Crystal structure of PBP3 from P. aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONZ
 
 | |
7ONK
 
 | | Crystal structure of PBP3 from P. aeruginosa in complex with AIC499 | | Descriptor: | (2S)-2-[(Z)-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2S)-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxy-3-[4-[N-[(3R)-piperidin-3-yl]carbamimidoyl]phenoxy]propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONX
 
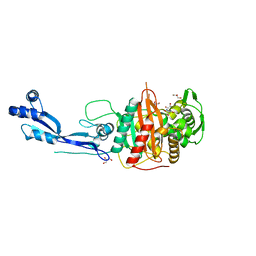 | | Crystal structure of PBP3 from P. aeruginosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
6DCJ
 
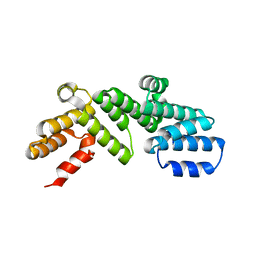 | |
4Y7P
 
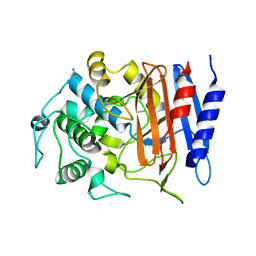 | | Structure of alkaline D-peptidase from Bacillus cereus | | Descriptor: | Alkaline D-peptidase, THIOCYANATE ION | | Authors: | Nakano, S, Okazaki, S, Ishitsubo, E, Kawahara, N, Komeda, H, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-02-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational analysis of peptide recognition mechanism of class-C type penicillin binding protein, alkaline D-peptidase from Bacillus cereus DF4-B
Sci Rep, 5, 2015
|
|
4L0L
 
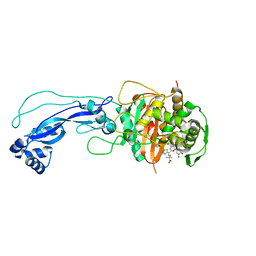 | | Crystal structure of P.aeruginosa PBP3 in complex with compound 4 | | Descriptor: | (6R,7S,10Z)-10-(2-amino-1,3-thiazol-4-yl)-1-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-7-formyl-13,13-dimethyl-3,9-dioxo-6-(sulfoamino)-12-oxa-2,4,8,11-tetraazatetradec-10-en-14-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S, Marr, E.S. | | Deposit date: | 2013-05-31 | | Release date: | 2013-08-21 | | Last modified: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyridone-conjugated monobactam antibiotics with gram-negative activity.
J.Med.Chem., 56, 2013
|
|
5FSR
 
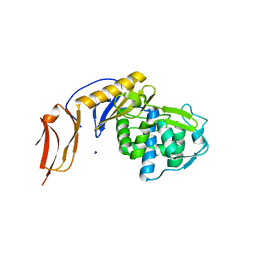 | | Crystal structure of penicillin binding protein 6B from Escherichia coli | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE DACD | | Authors: | Peters, K, Kannan, S, Rao, V.A, Bilboy, J, Vollmer, D, Erickson, S.W, Lewis, R.J, Young, K.D, Vollmer, W. | | Deposit date: | 2016-01-07 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Redundancy of Peptidoglycan Carboxypeptidases Ensures Robust Cell Shape Maintenance in Escherichia Coli
Mbio, 7, 2016
|
|
1MWT
 
 | |
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
