2I0T
 
 | |
2IAA
 
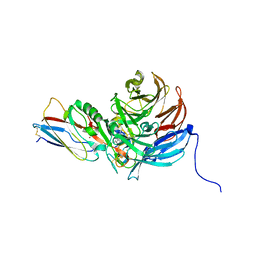 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 2) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-09-07 | | Release date: | 2006-11-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
2IUR
 
 | |
2IUP
 
 | |
1THM
 
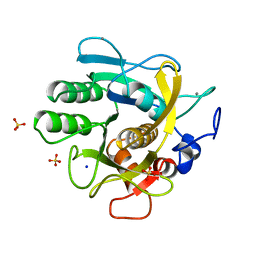 | | CRYSTAL STRUCTURE OF THERMITASE AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, SODIUM ION, SULFATE ION, ... | | Authors: | Teplyakov, A.V, Kuranova, I.P, Harutyunyan, E.H. | | Deposit date: | 1992-02-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of thermitase at 1.4 A resolution.
J.Mol.Biol., 214, 1990
|
|
2J57
 
 | | X-ray reduced Paraccocus denitrificans methylamine dehydrogenase N- quinol in complex with amicyanin. | | Descriptor: | AMICYANIN, COPPER (II) ION, METHYLAMINE DEHYDROGENASE HEAVY CHAIN, ... | | Authors: | Pearson, A.R, Pahl, R, Davidson, V.L, Wilmot, C.M. | | Deposit date: | 2006-09-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tracking X-Ray-Derived Redox Changes in Crystals of a Methylamine Dehydrogenase/Amicyanin Complex Using Single-Crystal Uv/Vis Microspectrophotometry.
J.Synchrotron Radiat., 14, 2007
|
|
4V62
 
 | | Crystal Structure of cyanobacterial Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Guskov, A, Gabdulkhakov, A, Kern, J, Broser, M, Zouni, A, Saenger, W. | | Deposit date: | 2008-01-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride
Nat.Struct.Mol.Biol., 16, 2009
|
|
4V82
 
 | | Crystal structure of cyanobacterial Photosystem II in complex with terbutryn | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gabdulkhakov, A, Broser, M, Guskov, A, Kern, J, Glockner, C, Muh, F, Saenger, W, Zouni, A. | | Deposit date: | 2010-11-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of cyanobacterial photosystem II Inhibition by the herbicide terbutryn
J.Biol.Chem., 286, 2011
|
|
3ZM8
 
 | | Crystal structure of Podospora anserina GH26-CBM35 beta-(1,4)- mannanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GH26 ENDO-BETA-1,4-MANNANASE, ... | | Authors: | Couturier, M, Roussel, A, Rosengren, A, Leone, P, Stalbrand, H, Berrin, J.G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 Beta-(1,4)-Mannanases from Podospora Anserina Reveal Differences Upon Manno-Oligosaccharides Catalysis.
J.Biol.Chem., 288, 2013
|
|
7W8U
 
 | | Crystal Structure of Indole Prenyltransferase IptA | | Descriptor: | 6-dimethylallyltryptophan synthase | | Authors: | Suemune, H, Nagano, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
6WT3
 
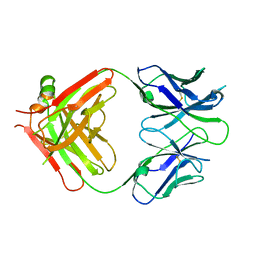 | | Structural basis for the binding of monoclonal antibody 5D2 to the tryptophan-rich lipid-binding loop in lipoprotein lipase | | Descriptor: | 5D2 FAB HEAVY CHAIN, 5D2 FAB LIGHT CHAIN | | Authors: | Luz, J.G, Birrane, G, Young, S.G, Meiyappan, M, Ploug, M. | | Deposit date: | 2020-05-01 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The structural basis for monoclonal antibody 5D2 binding to the tryptophan-rich loop of lipoprotein lipase.
J.Lipid Res., 61, 2020
|
|
6WN4
 
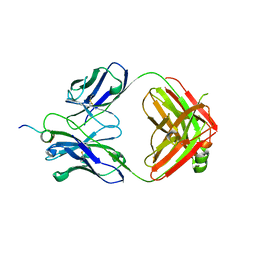 | | Structural basis for the binding of monoclonal antibody 5D2 to the tryptophan-rich lipid-binding loop in lipoprotein lipase | | Descriptor: | 5D2 FAB HEAVY CHAIN, 5D2 FAB LIGHT CHAIN, Lipoprotein lipase peptide | | Authors: | Luz, J.G, Birrane, G, Young, S.G, Meiyappan, M, Ploug, M. | | Deposit date: | 2020-04-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for monoclonal antibody 5D2 binding to the tryptophan-rich loop of lipoprotein lipase.
J.Lipid Res., 61, 2020
|
|
3E9G
 
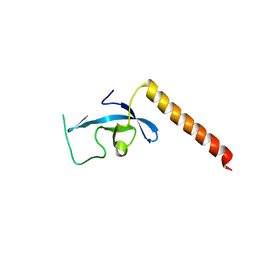 | | Crystal structure long-form (residue1-124) of Eaf3 chromo domain | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
6CDQ
 
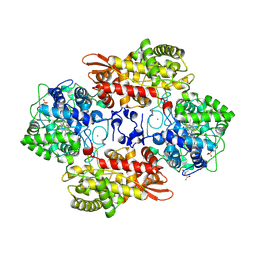 | |
5K6O
 
 | |
5K6M
 
 | |
5K6L
 
 | |
5KK9
 
 | | Connexin 32 G12R N-Terminal Mutant, | | Descriptor: | Gap junction beta-1 protein | | Authors: | Dowd, T.L, Barigello, T.A. | | Deposit date: | 2016-06-21 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Structural studies of N-terminal mutants of Connexin 26 and Connexin 32 using (1)H NMR spectroscopy.
Arch.Biochem.Biophys., 608, 2016
|
|
5D3U
 
 | | Crystal structure of the 5-selective H176F mutant of Cytochrome TxtE | | Descriptor: | CHLORIDE ION, GLYCEROL, P450-like protein, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Arnold, F.H. | | Deposit date: | 2015-08-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of a regioselectivity switch in nitrating P450s guided by molecular dynamics simulations and Markov models.
Nat.Chem., 8, 2016
|
|
5D40
 
 | | Crystal structure of the 5-selective H176Y mutant of Cytochrome TxtE | | Descriptor: | CHLORIDE ION, GLYCEROL, P450-like protein, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Arnold, F.H. | | Deposit date: | 2015-08-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Discovery of a regioselectivity switch in nitrating P450s guided by molecular dynamics simulations and Markov models.
Nat.Chem., 8, 2016
|
|
7BW0
 
 | | Active human TGR5 complex with a synthetic agonist 23H | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, G, Wang, X.K, Chen, Q, Hu, H.L, Ren, R.B. | | Deposit date: | 2020-04-12 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of activated bile acids receptor TGR5 in complex with stimulatory G protein.
Signal Transduct Target Ther, 5, 2020
|
|
7BZ2
 
 | | Cryo-EM structure of the formoterol-bound beta2 adrenergic receptor-Gs protein complex. | | Descriptor: | Beta2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, Y.N, Yang, F, Ling, S.L, Lv, P, Zhou, Y.X, Fang, W, Sun, W, Shi, P, Tian, C.L. | | Deposit date: | 2020-04-26 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Single-particle cryo-EM structural studies of the beta2AR-Gs complex bound with a full agonist formoterol.
Cell Discov, 6, 2020
|
|
2HXX
 
 | |
4R33
 
 | | X-ray structure of the tryptophan lyase NosL with Tryptophan and S-adenosyl-L-homocysteine bound | | Descriptor: | CHLORIDE ION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nicolet, Y, Zeppieri, L, Amara, P, Fontecilla-Camps, J.-C. | | Deposit date: | 2014-08-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Tryptophan Lyase (NosL): Evidence for Radical Formation at the Amino Group of Tryptophan.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4R34
 
 | | X-ray structure of the tryptophan lyase NosL with Tryptophan, 5'-deoxyadenosine and methionine bound | | Descriptor: | 5'-DEOXYADENOSINE, BROMIDE ION, GLYCEROL, ... | | Authors: | Nicolet, Y, Zeppieri, L, Amara, P, Fontecilla-Camps, J.-C. | | Deposit date: | 2014-08-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Tryptophan Lyase (NosL): Evidence for Radical Formation at the Amino Group of Tryptophan.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
