3CWV
 
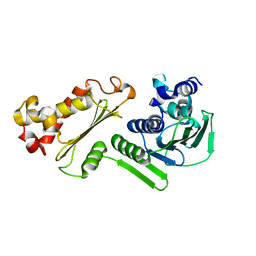 | | Crystal structure of B-subunit of the DNA gyrase from Myxococcus xanthus | | Descriptor: | DNA gyrase, B subunit, truncated | | Authors: | Ramagopal, U.A, Toro, R, Meyer, A.J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-22 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of B-subunit of the DNA gyrase from Myxococcus xanthus.
To be published
|
|
3COA
 
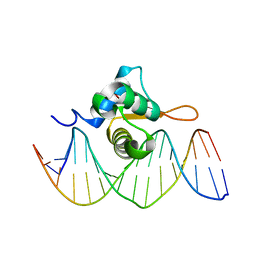 | | Crystal Structure of FoxO1 DBD Bound to IRE DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*DCP*DAP*DAP*DGP*DCP*DAP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DCP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DGP*DTP*DTP*DTP*DGP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DG)-3'), ... | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
3CON
 
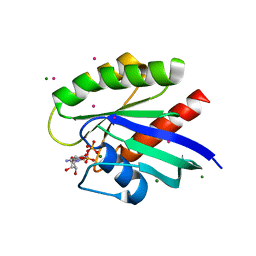 | | Crystal structure of the human NRAS GTPase bound with GDP | | Descriptor: | GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Shen, L, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal structure of the human NRAS GTPase bound with GDP.
To be Published
|
|
3CZL
 
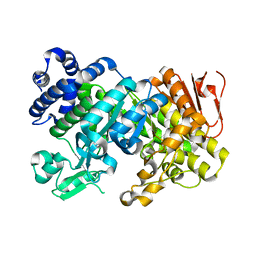 | |
2RBD
 
 | |
3D1G
 
 | | Structure of a small molecule inhibitor bound to a DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, [(5R)-5-(2,3-dibromo-5-ethoxy-4-hydroxybenzyl)-4-oxo-2-thioxo-1,3-thiazolidin-3-yl]acetic acid | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3D22
 
 | | Crystal structure of a poplar thioredoxin h mutant, PtTrxh4C61S | | Descriptor: | PHOSPHATE ION, Thioredoxin H-type | | Authors: | Koh, C.S, Didierjean, C, Corbier, C, Rouhier, N, Jacquot, J.P, Gelhaye, E. | | Deposit date: | 2008-05-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Atypical Catalytic Mechanism Involving Three Cysteines of Thioredoxin.
J.Biol.Chem., 283, 2008
|
|
2RCC
 
 | |
3D6I
 
 | | Structure of the Thioredoxin-like Domain of Yeast Glutaredoxin 3 | | Descriptor: | Monothiol glutaredoxin-3, SULFATE ION | | Authors: | Lebioda, L, Gibson, L.M, Dingra, N.N, Outten, C.E. | | Deposit date: | 2008-05-19 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the thioredoxin-like domain of yeast glutaredoxin 3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
5Z0X
 
 | |
3CED
 
 | | Crystal structure of the C-terminal NIL domain of an ABC transporter protein homologue from Staphylococcus aureus | | Descriptor: | 1,4-BUTANEDIOL, Methionine import ATP-binding protein metN 2 | | Authors: | Cuff, M.E, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-28 | | Release date: | 2008-05-13 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of the C-terminal NIL domain of an ABC transporter protein homologue from Staphylococcus aureus.
TO BE PUBLISHED
|
|
2RH0
 
 | |
2RIL
 
 | |
3CUP
 
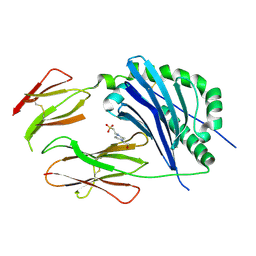 | | Crystal structure of the MHC class II molecule I-Ag7 in complex with the peptide GAD221-235 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, H-2 class II histocompatibility antigen, ... | | Authors: | Corper, A.L, Yoshida, K, Teyton, L, Wilson, I.A. | | Deposit date: | 2008-04-16 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of the MHC class II molecule I-Ag7 in complex with the peptide GAD221-235
To be Published
|
|
3CNG
 
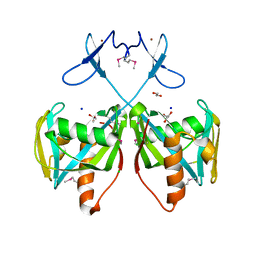 | | Crystal structure of NUDIX hydrolase from Nitrosomonas europaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
3CO6
 
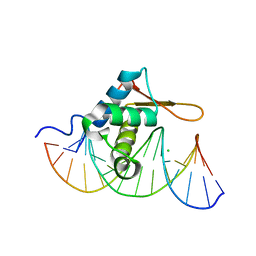 | | Crystal Structure of FoxO1 DBD Bound to DBE1 DNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DCP*DAP*DAP*DGP*DGP*DTP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DCP*DCP*DA)-3'), ... | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
3CQW
 
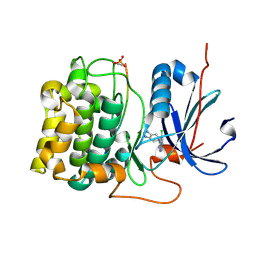 | | Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor | | Descriptor: | 5-(5-chloro-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine, Glycogen synthase kinase-3 beta, MANGANESE (II) ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure based optimization of novel Akt inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CTY
 
 | |
2RAU
 
 | |
3D3P
 
 | |
3CW7
 
 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Cytosine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(8OG)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CWT
 
 | | Crystal Structure of an AlkA Host/Guest Complex 2'-fluoro-2'-deoxyinosine:Adenine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(2FI)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DAP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CXS
 
 | | Crystal structure of human GNA1 | | Descriptor: | Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
2RE3
 
 | |
