5ZH0
 
 | | Crystal Structures of Endo-beta-1,4-xylanase II | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | Authors: | Zhang, X, Wan, Q, Li, Z. | | Deposit date: | 2018-03-10 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal Structures of Endo-beta-1,4-xylanase II
To be published
|
|
5OL4
 
 | | 1.28 A resolution of Sporosarcina pasteurii urease inhibited in the presence of NBPT | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Phosphoramidothioic O,O-acid, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2017-07-26 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Urease Inhibition in the Presence of N-(n-Butyl)thiophosphoric Triamide, a Suicide Substrate: Structure and Kinetics.
Biochemistry, 56, 2017
|
|
5TUQ
 
 | | Crystal Structure of a 6-Cyclohexylmethyl-3-hydroxypyrimidine-2,4-dione Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 1-[(benzyloxy)methyl]-6-(cyclohexylmethyl)-3-hydroxy-5-(propan-2-yl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | 6-Cyclohexylmethyl-3-hydroxypyrimidine-2,4-dione as an inhibitor scaffold of HIV reverase transcriptase: Impacts of the 3-OH on inhibiting RNase H and polymerase.
Eur J Med Chem, 128, 2017
|
|
7FIP
 
 | | The native structure of beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, ZINC ION | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
4JB7
 
 | | 1.42 Angstrom resolution crystal structure of accessory colonization factor AcfC (acfC) in complex with D-aspartic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Accessory colonization factor AcfC, D-MALATE, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Dubrovska, I, Winsor, J, Minasov, G, Shuvalova, L, Filippova, E.V, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42 Angstrom resolution crystal structure of accessory colonization factor AcfC (acfC) in complex with D-aspartic acid
To be Published
|
|
3MST
 
 | |
3HB4
 
 | | 17beta-hydroxysteroid dehydrogenase type1 complexed with E2B | | Descriptor: | 3-{[(9beta,14beta,16alpha,17alpha)-3,17-dihydroxyestra-1,3,5(10)-trien-16-yl]methyl}benzamide, Estradiol 17-beta-dehydrogenase 1 | | Authors: | Mazumdar, M, Fournier, D, Zhu, D.-W, Cadot, C, Poirier, D, Lin, S.-X. | | Deposit date: | 2009-05-04 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Binary and ternary crystal structure analyses of a novel inhibitor with 17beta-HSD type 1: a lead compound for breast cancer therapy.
Biochem.J., 424, 2009
|
|
3TGY
 
 | | Crystal structure of the complex of Bovine Lactoperoxidase with Ascorbic acid at 2.35 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Singh, R.P, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-08-18 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution.
Biochim.Biophys.Acta, 1865, 2017
|
|
5YPN
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI2 complex | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Liu, W, Wang, D. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
3V66
 
 | | HUMAN SQUALENE SYNTHASE IN COMPLEX WITH 2-(1-{2-[(4R,6S)-8-chloro-6-(2,3-dimethoxyphenyl)-4H,6H-pyrrolo[1,2-a][4,1]benzoxazepin-4-yl]acetyl}-4-piperidinyl)acetic acid | | Descriptor: | (1-{[(4R,6S)-8-chloro-6-(2,3-dimethoxyphenyl)-4H,6H-pyrrolo[1,2-a][4,1]benzoxazepin-4-yl]acetyl}piperidin-4-yl)acetic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Ohtsuka, M, Ohki, H, Haginoya, N, Itoh, M, Sugita, K, Usui, H, Ichikawa, M, Higashihashi, N. | | Deposit date: | 2011-12-18 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of novel tricyclic compounds as squalene synthase inhibitors
Bioorg.Med.Chem., 20, 2012
|
|
2X2H
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase | | Descriptor: | ACETATE ION, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
3SZ9
 
 | |
3RQT
 
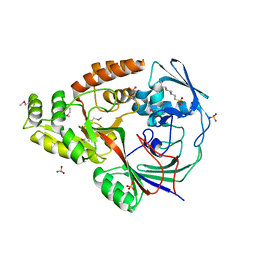 | | 1.5 Angstrom Crystal Structure of the Complex of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Nickel and two Histidines | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HISTIDINE, NICKEL (II) ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Falugi, F, Bottomley, M, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of the Complex of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Nickel and two Histidines.
TO BE PUBLISHED
|
|
5UFN
 
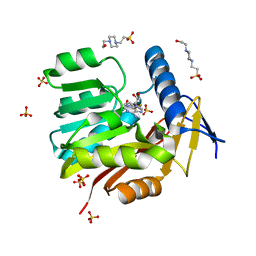 | | Crystal structure of Burkholderia thailandensis 1,6-didemethyltoxoflavin-N1-methyltransferase with bound S-adenosylhomocysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Methyltransferase domain protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fenwick, M.K, Ealick, S.E, Almabruk, K.H, Begley, T.P, Philmus, B. | | Deposit date: | 2017-01-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Biochemical Characterization and Structural Basis of Reactivity and Regioselectivity Differences between Burkholderia thailandensis and Burkholderia glumae 1,6-Didesmethyltoxoflavin N-Methyltransferase.
Biochemistry, 56, 2017
|
|
6F9U
 
 | |
3K9Q
 
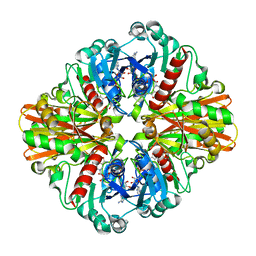 | | Crystal structure of C151G mutant of Glyceraldehyde 3-phosphate dehydrogenase 1 from Methicillin resistant Staphylococcus aureus (MRSA252) at 2.5 angstrom resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase 1, ... | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-10-16 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
7Y5K
 
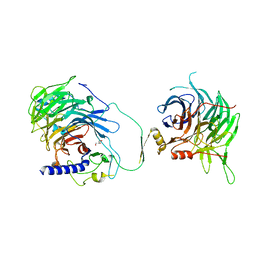 | | Crystal structure of human CAF-1 core complex in spacegroup C2221 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
4CLH
 
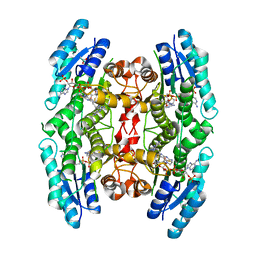 | |
4CME
 
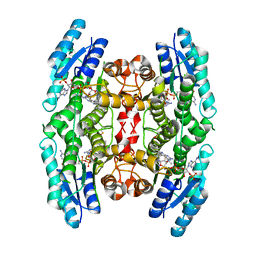 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | N4,N4-dimethyl-5,6-diphenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 1 | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CJX
 
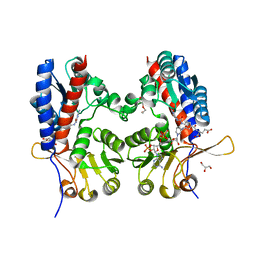 | | The crystal structure of Trypanosoma brucei N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) complexed with NADP cofactor and inhibitor | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC, ... | | Authors: | Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2013-12-23 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of 2,4-Diamino-6-Oxo-1,6-Dihydropyrimidin-5-Yl Ureido Based Inhibitors of Trypanosoma Brucei Fold and Testing for Antiparasitic Activity.
J.Med.Chem., 58, 2015
|
|
6JRX
 
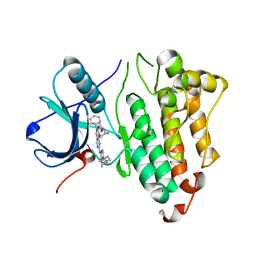 | | EGFR T790M/C797S in complex with compound 6i | | Descriptor: | Epidermal growth factor receptor, N-{trans-4-[3-(2-chlorophenyl)-7-{[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]cyclohexyl}propanamide | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JIZ
 
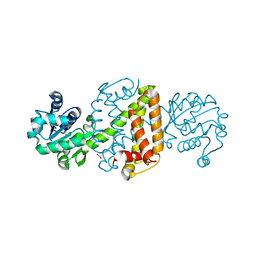 | | Apo structure of an imine reductase at 1.76 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-ethylheptane, 6-phosphogluconate dehydrogenase NAD-binding protein, ... | | Authors: | Li, H, Wu, L, Zheng, G.W, Zhou, J.H. | | Deposit date: | 2019-02-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Apo structure of an imine reductase at 1.76 Angstrom resolution
To Be Published
|
|
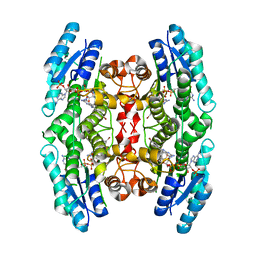
4CMJ
 
 | |
1QUS
 
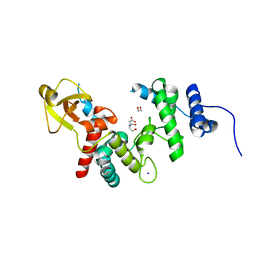 | | 1.7 A RESOLUTION STRUCTURE OF THE SOLUBLE LYTIC TRANSGLYCOSYLASE SLT35 FROM ESCHERICHIA COLI | | Descriptor: | 1,2-ETHANEDIOL, BICINE, LYTIC MUREIN TRANSGLYCOSYLASE B, ... | | Authors: | van Asselt, E.J, Dijkstra, A.J, Kalk, K.H, Takacs, B, Keck, W, Dijkstra, B.W. | | Deposit date: | 1999-07-03 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand.
Structure Fold.Des., 7, 1999
|
|
3JS8
 
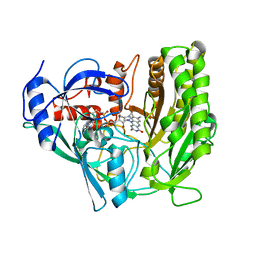 | | Solvent-stable cholesterol oxidase | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Sagermann, M, Ohtaki, A, Newton, K, Doukyu, N. | | Deposit date: | 2009-09-09 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural characterization of the organic solvent-stable cholesterol oxidase from Chromobacterium sp. DS-1.
J.Struct.Biol., 170, 2010
|
|
