6TNN
 
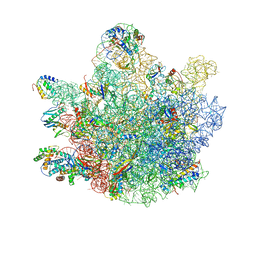 | | Mini-RNase III (Mini-III) bound to 50S ribosome with precursor 23S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
5DGV
 
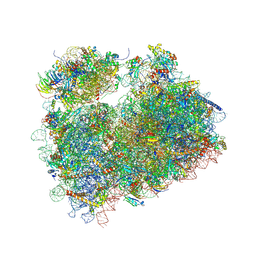 | | Complex of yeast 80S ribosome with hypusine-containing/non-modified eIF5A and/or a peptidyl-tRNA analog | | Descriptor: | 25S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-28 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
4U50
 
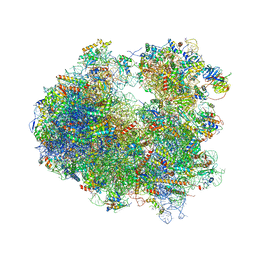 | | Crystal structure of Verrucarin bound to the yeast 80S ribosome | | Descriptor: | (4S,5R,10E,12Z,16R,16aS,17S,18R,19aR,23aR)-4-hydroxy-5,16a,21-trimethyl-4,5,6,7,16,16a,22,23-octahydro-3H,18H,19aH-spiro[16,18-methano[1,6,12]trioxacyclooctadecino[3,4-d]chromene-17,2'-oxirane]-3,9,14-trione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
6CAQ
 
 | |
5MMM
 
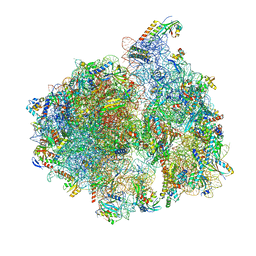 | | Structure of the 70S chloroplast ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein 2, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
5GMF
 
 | | Crystal structure of monkey TLR7 in complex with guanosine and polyU | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA
Immunity, 45, 2016
|
|
5IB8
 
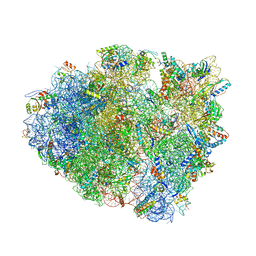 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and near-cognate tRNALys with U-G mismatch in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
5A03
 
 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with xylose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ALDOSE-ALDOSE OXIDOREDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
4U8I
 
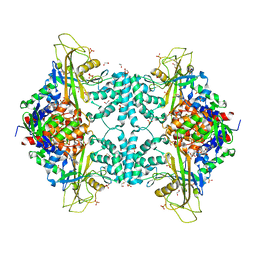 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
5CZ8
 
 | |
5MKL
 
 | |
6CB3
 
 | |
4U3U
 
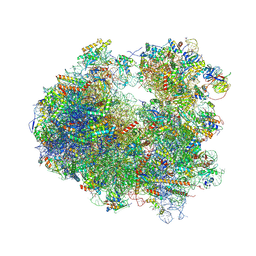 | | Crystal structure of Cycloheximide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
5D0Z
 
 | |
5DA5
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 | | Descriptor: | CALCIUM ION, FE (III) ION, GLYCOLIC ACID, ... | | Authors: | He, D, Vanden Hehier, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5DBU
 
 | |
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
4U4Z
 
 | | Crystal structure of Phyllanthoside bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U6F
 
 | | Crystal structure of T-2 toxin bound to the yeast 80S ribosome | | Descriptor: | 12,13-Epoxytrichothec-9-ene-3,4,8,15-tetrol-4,15-diacetate-8-isovalerate, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-28 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U8K
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Q107A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
5M73
 
 | | Structure of the human SRP S domain with SRP72 RNA-binding domain | | Descriptor: | GLYCEROL, Human gene for small cytoplasmic 7SL RNA (7L30.1), MAGNESIUM ION, ... | | Authors: | Becker, M.M.M, Wild, K, Sinning, I. | | Deposit date: | 2016-10-26 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human SRP72 complexes provide insights into SRP RNA remodeling and ribosome interaction.
Nucleic Acids Res., 45, 2017
|
|
4ZUX
 
 | |
5MP9
 
 | | 26S proteasome in presence of ATP (s1) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6TIX
 
 | |
6T9N
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
