1MCO
 
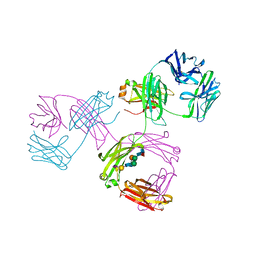 | | THREE-DIMENSIONAL STRUCTURE OF A HUMAN IMMUNOGLOBULIN WITH A HINGE DELETION | | Descriptor: | IGG1 MCG INTACT ANTIBODY (HEAVY CHAIN), IGG1 MCG INTACT ANTIBODY (LIGHT CHAIN), N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-L-gulopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guddat, L.W, Edmundson, A.B. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Three-dimensional structure of a human immunoglobulin with a hinge deletion.
Proc.Natl.Acad.Sci.Usa, 90, 1993
|
|
5UIX
 
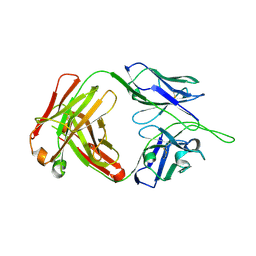 | |
4JY2
 
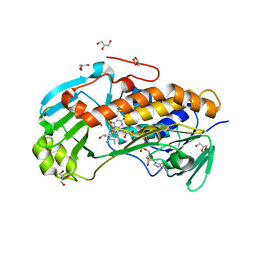 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, native and unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
2FUA
 
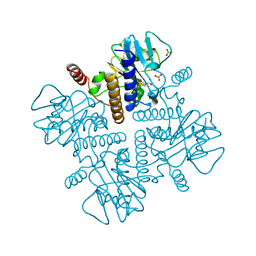 | | L-FUCULOSE 1-PHOSPHATE ALDOLASE CRYSTAL FORM T WITH COBALT | | Descriptor: | BETA-MERCAPTOETHANOL, COBALT (II) ION, L-FUCULOSE-1-PHOSPHATE ALDOLASE, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined high-resolution structure of the metal-ion dependent L-fuculose-1-phosphate aldolase (class II) from Escherichia coli.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2GEZ
 
 | | Crystal structure of potassium-independent plant asparaginase | | Descriptor: | CHLORIDE ION, L-asparaginase alpha subunit, L-asparaginase beta subunit, ... | | Authors: | Michalska, K, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of plant asparaginase.
J.Mol.Biol., 360, 2006
|
|
2GHB
 
 | | Thermotoga maritima maltotriose binding protein, ligand free form | | Descriptor: | maltose ABC transporter, periplasmic maltose-binding protein | | Authors: | Cuneo, M.J, Changela, A, Hocker, B, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2006-03-27 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | T. maritima maltotriose binding protein open form
To be Published
|
|
1BT7
 
 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | Descriptor: | NS3 SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
1SC6
 
 | |
3BCP
 
 | | Crystal Structure of The Swapped non covalent form of P19A/L28Q/N67D BS-RNase | | Descriptor: | Seminal ribonuclease | | Authors: | Merlino, A, Ercole, C, Picone, D, Pizzo, E, Mazzarella, L, Sica, F. | | Deposit date: | 2007-11-13 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The buried diversity of bovine seminal ribonuclease: shape and cytotoxicity of the swapped non-covalent form of the enzyme
J.Mol.Biol., 376, 2008
|
|
1O8B
 
 | | Structure of Escherichia coli ribose-5-phosphate isomerase, RpiA, complexed with arabinose-5-phosphate. | | Descriptor: | 5-O-phosphono-beta-D-arabinofuranose, RIBOSE 5-PHOSPHATE ISOMERASE | | Authors: | Zhang, R.-g, Andersson, C.E, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Mowbray, S.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-11-26 | | Release date: | 2003-01-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of Escherichia Coli Ribose-5-Phosphate Isomerase: A Ubiquitous Enzyme of the Pentose Phosphate Pathway and the Calvin Cycle
Structure, 11, 2003
|
|
1CAK
 
 | | STRUCTURAL ANALYSIS OF THE ZINC HYDROXIDE-THR 199-GLU 106 HYDROGEN BONDING NETWORK IN HUMAN CARBONIC ANHYDRASE II | | Descriptor: | CARBONIC ANHYDRASE II, SULFATE ION, ZINC ION | | Authors: | Xue, Y, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II.
Proteins, 17, 1993
|
|
1NTP
 
 | |
1PN3
 
 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and the acceptor substrate DVV. | | Descriptor: | DESVANCOSAMINYL VANCOMYCIN, GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-06-12 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4JJ9
 
 | | Crystal Structure of 5-carboxymethyl-2-hydroxymuconate delta-isomerase | | Descriptor: | 5-carboxymethyl-2-hydroxymuconate delta-isomerase, SULFATE ION | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-07 | | Release date: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of 5-carboxymethyl-2-hydroxymuconate delta-isomerase
To be Published
|
|
5U15
 
 | |
1D2Q
 
 | | CRYSTAL STRUCTURE OF HUMAN TRAIL | | Descriptor: | TNF-RELATED APOPTOSIS INDUCING LIGAND | | Authors: | Cha, S.-S. | | Deposit date: | 1999-09-27 | | Release date: | 2000-02-11 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 A resolution crystal structure of human TRAIL, a cytokine with selective antitumor activity.
Immunity, 11, 1999
|
|
2QYN
 
 | | Crystal structure of PDE4D2 in complex with inhibitor NPV | | Descriptor: | 4-[8-(3-nitrophenyl)-1,7-naphthyridin-6-yl]benzoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ke, H. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
1R3M
 
 | | Crystal structure of the dimeric unswapped form of bovine seminal ribonuclease | | Descriptor: | PHOSPHATE ION, Ribonuclease, seminal | | Authors: | Berisio, R, Sica, F, De Lorenzo, C, Di Fiore, A, Piccoli, R, Zagari, A, Mazzarella, L. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the dimeric unswapped form of bovine seminal ribonuclease
Febs Lett., 554, 2003
|
|
1MA9
 
 | | Crystal structure of the complex of human vitamin D binding protein and rabbit muscle actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Alpha Skeletal Muscle, ... | | Authors: | Verboven, C, Bogaerts, I, Waelkens, E, Rabijns, A, Van Baelen, H, Bouillon, R, De Ranter, C. | | Deposit date: | 2002-08-02 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Actin-DBP: the perfect structural fit?
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MDO
 
 | | Crystal structure of ArnB aminotransferase with pyridomine 5' phosphate | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ArnB aminotransferase | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
7K2U
 
 | | DHODH IN COMPLEX WITH LIGAND 13 | | Descriptor: | 6-fluoro-2-(2'-fluoro[1,1'-biphenyl]-4-yl)-N-methoxy-3-methylquinoline-4-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2020-09-09 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A carboxylic acid isostere screen of the DHODH inhibitor Brequinar.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
1ULV
 
 | | Crystal Structure of Glucodextranase Complexed with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
1UVF
 
 | |
2F8C
 
 | | Crystal structure of FPPS in complex with Zoledronate | | Descriptor: | Farnesyl Diphosphate Synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rondeau, J.-M, Bitsch, F, Bourgier, E, Geiser, M, Hemmig, R, Kroemer, M, Lehmann, S, Ramage, P, Rieffel, S, Strauss, A, Green, J.R, Jahnke, W. | | Deposit date: | 2005-12-02 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs.
Chemmedchem, 1, 2006
|
|
2F92
 
 | | Crystal structure of human FPPS in complex with alendronate | | Descriptor: | 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, Farnesyl Diphosphate Synthase, PHOSPHATE ION, ... | | Authors: | Rondeau, J.-M, Bitsch, F, Bourgier, E, Geiser, M, Hemmig, R, Kroemer, M, Lehmann, S, Ramage, P, Rieffel, S, Strauss, A, Green, J.R, Jahnke, W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs.
Chemmedchem, 1, 2006
|
|
