1BLK
 
 | | NMR ENSEMBLE OF BLK SH2 DOMAIN USING CHEMICAL SHIFT REFINEMENT, 20 STRUCTURES | | Descriptor: | P55 BLK PROTEIN TYROSINE KINASE | | Authors: | Metzler, W.J, Leiting, B, Pryor, K, Mueller, L, Farmer II, B.T. | | Deposit date: | 1996-03-26 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the SH2 domain from p55blk kinase.
Biochemistry, 35, 1996
|
|
1CQ5
 
 | | NMR STRUCTURE OF SRP RNA DOMAIN IV | | Descriptor: | SRP RNA DOMAIN IV | | Authors: | Schmitz, U, James, T.L, Behrens, S, Freymann, D.M, Lukavsky, P, Walter, P. | | Deposit date: | 1999-08-05 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the phylogenetically most conserved domain of SRP RNA.
RNA, 5, 1999
|
|
1CMR
 
 | | NMR SOLUTION STRUCTURE OF A CHIMERIC PROTEIN, DESIGNED BY TRANSFERRING A FUNCTIONAL SNAKE BETA-HAIRPIN INTO A SCORPION ALPHA/BETA SCAFFOLD (PH 3.5, 20C), NMR, 18 STRUCTURES | | Descriptor: | CHARYBDOTOXIN, ALPHA CHIMERA | | Authors: | Zinn-Justin, S, Guenneugues, M, Drakopoulou, E, Gilquin, B, Vita, C, Menez, A. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Transfer of a beta-hairpin from the functional site of snake curaremimetic toxins to the alpha/beta scaffold of scorpion toxins: three-dimensional solution structure of the chimeric protein.
Biochemistry, 35, 1996
|
|
2RRS
 
 | |
5J3F
 
 | | NMR solution structure of [Rp, Rp]-PT dsDNA | | Descriptor: | DNA (5'-D(*CP*GP*(RSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(RSG)P*CP*CP*G)-3') | | Authors: | Lan, W, Hu, Z, Cao, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
5J3I
 
 | | NMR solution structure of [Sp, Sp]-PT dsDNA | | Descriptor: | DNA (5'-D(*CP*GP*(SSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(SSG)P*CP*CP*G)-3') | | Authors: | Lan, W, Hu, Z, Cao, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
7L8V
 
 | |
6Q2Z
 
 | | NMR solution structure of the HVO_2922 protein from Haloferax volcanii | | Descriptor: | UPF0339 family protein | | Authors: | Kubatova, N, Jonker, H.R.A, Saxena, K, Richter, C, Marchfelder, A, Schwalbe, H. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Small Protein HVO_2922 from Haloferax volcanii.
Chembiochem, 21, 2020
|
|
6KFI
 
 | | NMR solution structure of the 1:1 complex of Tel26 G-quadruplex and a tripodal cationic fluorescent probe NBTE | | Descriptor: | 4,4',4''-(nitrilotris(benzene-4,1-diyl))tris(1-ethylpyridin-1-ium) iodide, G-quadruplex DNA (26-MER) | | Authors: | Liu, W, Liu, L.Y, Mao, Z.W. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6KFJ
 
 | | NMR solution structure of the 1:1 complex of wtTel26 G-quadruplex and a tripodal cationic fluorescent probe NBTE | | Descriptor: | 4,4',4''-(nitrilotris(benzene-4,1-diyl))tris(1-ethylpyridin-1-ium) iodide, G-quadruplex DNA wtTel26 | | Authors: | Liu, W, Liu, L.Y, Mao, Z.W. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6QS0
 
 | |
5UKZ
 
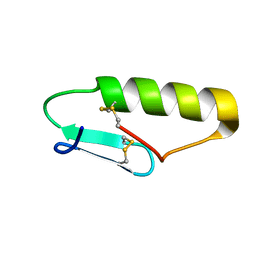 | | NMR Solution structure of chemically synthesized antilisterial Pediocin PA-1 M31L analog. | | Descriptor: | Bacteriocin pediocin PA-1 M31L | | Authors: | Bedard, F, Hammami, R, Zirah, S, Rebuffat, S, Fliss, I, Biron, E. | | Deposit date: | 2017-01-23 | | Release date: | 2018-06-27 | | Method: | SOLUTION NMR | | Cite: | Synthesis, antimicrobial activity and conformational analysis of the class IIa bacteriocin pediocin PA-1 and analogs thereof.
Sci Rep, 8, 2018
|
|
6DO7
 
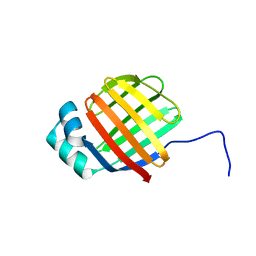 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
6G04
 
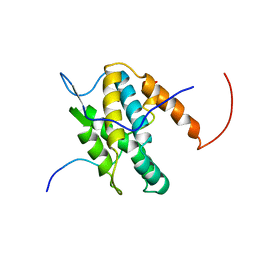 | | NMR Solution Structure of Yeast TSR2(1-152) in Complex with S26A(100-119) | | Descriptor: | 40S ribosomal protein S26-A, Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
6QKP
 
 | |
6QKQ
 
 | |
6SGO
 
 | | NMR structure of MLP124017 | | Descriptor: | Secreted protein | | Authors: | Barthe, P, de Guillen, K, Padilla, A, Hecker, A. | | Deposit date: | 2019-08-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural genomics applied to the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine knot and NTF2-like protein folds.
Sci Rep, 9, 2019
|
|
6G03
 
 | | NMR Solution Structure of yeast TSR2(1-152) | | Descriptor: | Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
5LFH
 
 | | NMR structure of peptide 10 targeting CXCR4 | | Descriptor: | ACE-ARG-ALA-DCY-ARG-PHE-PHE-CYS | | Authors: | Di Maro, S, Trotta, A.M, Brancaccio, D, Di Leva, F.S, La Pietra, V, Ierano, C, Napolitano, M, Portella, L, D'Alterio, C, Siciliano, R.A, Sementa, D, Tomassi, S, Carotenuto, A, Novellino, E, Scala, S, Marinelli, L. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2016-10-05 | | Method: | SOLUTION NMR | | Cite: | Exploring the N-Terminal Region of C-X-C Motif Chemokine 12 (CXCL12): Identification of Plasma-Stable Cyclic Peptides As Novel, Potent C-X-C Chemokine Receptor Type 4 (CXCR4) Antagonists.
J.Med.Chem., 59, 2016
|
|
5LM0
 
 | | NMR spatial structure of Tk-hefu peptide | | Descriptor: | L-2 | | Authors: | Mineev, K.S, Berkut, A.A, Novikova, E.V, Oparin, P.B, Grishin, E.V, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein Surface Topography as a tool to enhance the selective activity of a potassium channel blocker.
J.Biol.Chem., 2019
|
|
5LFF
 
 | | NMR structure of peptide 2 targeting CXCR4 | | Descriptor: | ARG-ALA-CYS-ARG-PHE-PHE-CYS | | Authors: | Di Maro, S, Trotta, A.M, Brancaccio, D, Di Leva, F.S, La Pietra, V, Ierano, C, Napolitano, M, Portella, L, D'Alterio, C, Siciliano, R.A, Sementa, D, Tomassi, S, Carotenuto, A, Novellino, E, Scala, S, Marinelli, L. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Exploring the N-Terminal Region of C-X-C Motif Chemokine 12 (CXCL12): Identification of Plasma-Stable Cyclic Peptides As Novel, Potent C-X-C Chemokine Receptor Type 4 (CXCR4) Antagonists.
J.Med.Chem., 59, 2016
|
|
5LAH
 
 | | NMR structure of the sea anemone peptide tau-AnmTx Ueq 12-1 with an uncommon fold | | Descriptor: | tau-AnmTx Ueq 12-1 | | Authors: | Mineev, K.S, Arseniev, A.S, Andreev, Y.A, Kozlov, S.A, Logashina, Y.A. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Disulfide-Stabilized Fold Provides Sea Anemone Peptide to Exhibit Both Antimicrobial and TRPA1 Potentiating Properties
Toxins, 9, 2017
|
|
6GT7
 
 | |
6HT4
 
 | |
1CTO
 
 | | NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF THE LIGAND-BINDING REGION OF MURINE GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR | | Authors: | Yamasaki, K, Naito, S, Anaguchi, H, Ohkubo, T, Ota, Y. | | Deposit date: | 1996-09-25 | | Release date: | 1997-10-22 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an extracellular domain containing the WSxWS motif of the granulocyte colony-stimulating factor receptor and its interaction with ligand.
Nat.Struct.Biol., 4, 1997
|
|
