7N17
 
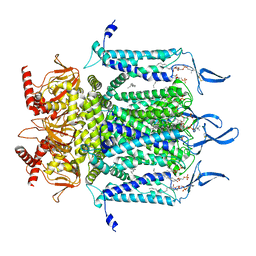 | | Structure of TAX-4_R421W apo open state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
5WDT
 
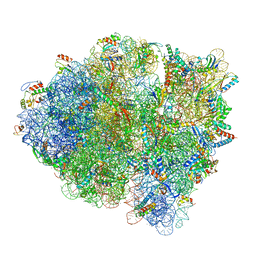 | | 70S ribosome-EF-Tu H84A complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-06 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
7N16
 
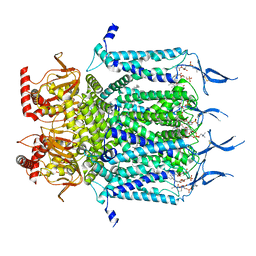 | | Structure of TAX-4_R421W apo closed state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7EMK
 
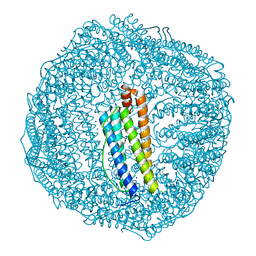 | | Dendrorhynchus zhejiangensis ferritin | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dendrorhynchus zhejiangensis ferritin, ... | | Authors: | Huan, H.S, Ming, T.H, Su, X.R. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of ferritin from Dendrorhynchus zhejiangensis
Biochemical and Biophysical Research Communications, 531, 2020
|
|
5TBW
 
 | | Crystal structure of chlorolissoclimide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Konst, Z.A, Szklarski, A.R, Pellegrino, S, Michalak, S.E, Meyer, M, Zanette, C, Cencic, R, Nam, S, Horne, D.A, Pelletier, J, Mobley, D.L, Yusupova, G, Yusupov, M, Vanderwal, C.D. | | Deposit date: | 2016-09-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides.
Nat Chem, 9, 2017
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEK
 
 | | Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2BKC
 
 | |
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5WE4
 
 | | 70S ribosome-EF-Tu wt complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
5VK0
 
 | |
8SF7
 
 | | 48-nm doublet microtubule from Tetrahymena thermophila strain MEC17 | | Descriptor: | CFAM166A, CFAM166B, CFAM166C, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Effect of alpha-tubulin acetylation on the doublet microtubule structure.
Elife, 12, 2024
|
|
4Y18
 
 | |
5KK3
 
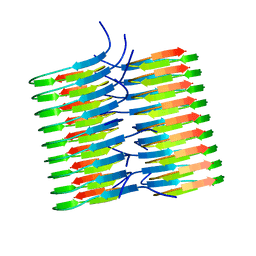 | | Atomic Resolution Structure of Monomorphic AB42 Amyloid Fibrils | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Colvin, M.T, Silvers, R, Zhe Ni, Q, Can, T.V, Sergeyev, I, Rosay, M, Donovan, K.J, Michael, B, Wall, J, Linse, S, Griffin, R.G. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic Resolution Structure of Monomorphic A beta 42 Amyloid Fibrils.
J.Am.Chem.Soc., 138, 2016
|
|
4P6F
 
 | |
2GTH
 
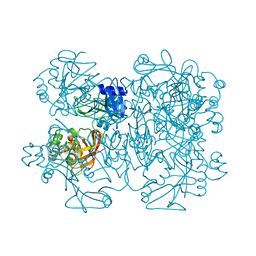 | | crystal structure of the wildtype MHV coronavirus non-structural protein nsp15 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Xu, X, Zhai, Y, Sun, F, Lou, Z, Su, D, Rao, Z. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New Antiviral Target Revealed by the Hexameric Structure of Mouse Hepatitis Virus Nonstructural Protein nsp15
J.Virol., 80, 2006
|
|
2GTI
 
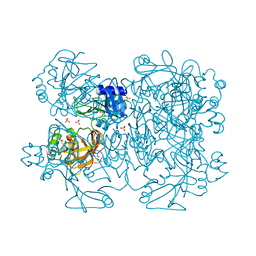 | | mutation of MHV coronavirus non-structural protein nsp15 (F307L) | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, SULFATE ION | | Authors: | Xu, X, Zhai, Y, Sun, F, Lou, Z, Su, D, Rao, Z. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-15 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | New Antiviral Target Revealed by the Hexameric Structure of Mouse Hepatitis Virus Nonstructural Protein nsp15
J.Virol., 80, 2006
|
|
2EWO
 
 | |
8IQH
 
 | |
8IQI
 
 | | Structure of Full-Length AsfvPrimPol in Complex-Form | | Descriptor: | DNA (32-MER), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Shao, Z.W, Su, S.C, Gan, J.H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures and implications of the C962R protein of African swine fever virus.
Nucleic Acids Res., 51, 2023
|
|
7DT1
 
 | | The structure of Lactobacillus fermentum 4,6-alpha-Glucanotransferase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W.K, Yong, Y.H, Wu, L, Chen, S, Zhou, J.H, Wu, J. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43002272 Å) | | Cite: | Characterization of a new 4,6-alpha-glucanotransferase from Limosilactobacillus fermentum NCC 3057 with ability of synthesizing low molecular mass isomalto-/maltopolysaccharide
Food Biosci, 46, 2022
|
|
7SFB
 
 | |
3U13
 
 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
2QOP
 
 | | Crystal structure of the transcriptional regulator AcrR from Escherichia coli | | Descriptor: | HTH-type transcriptional regulator acrR | | Authors: | Li, M, Gu, R, Su, C.-C, McDermott, G, Yu, E.W. | | Deposit date: | 2007-07-20 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the transcriptional regulator AcrR from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
7SGH
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML124N | | Descriptor: | (S)-N-((S)-1-imino-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-4-methyl-2-(2-((2,4,6-trifluorophenyl)amino)acetamido)pentanamide, 3C-like proteinase | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
