1DM9
 
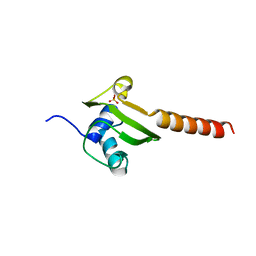 | | HEAT SHOCK PROTEIN 15 KD | | Descriptor: | HYPOTHETICAL 15.5 KD PROTEIN IN MRCA-PCKA INTERGENIC REGION, SULFATE ION | | Authors: | Staker, B.L, Korber, P, Bardwell, J.C.A, Saper, M.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hsp15 reveals a novel RNA-binding motif.
EMBO J., 19, 2000
|
|
1DMG
 
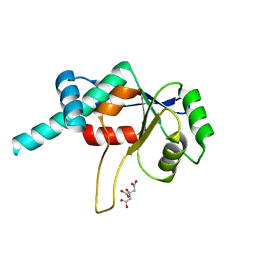 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L4 | | Descriptor: | CITRIC ACID, RIBOSOMAL PROTEIN L4 | | Authors: | Worbs, M, Huber, R, Wahl, M.C. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of ribosomal protein L4 shows RNA-binding sites for ribosome incorporation and feedback control of the S10 operon.
EMBO J., 19, 2000
|
|
7WMH
 
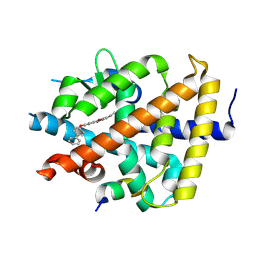 | | A novel chemical derivative(56) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-(4-phenylphenoxy)isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMG
 
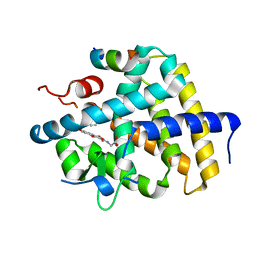 | | A novel chemical derivative(52) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-(4-phenoxyphenoxy)isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMJ
 
 | | A novel chemical derivative(71) of THRB agonist | | Descriptor: | 2-[[7-[2,6-dimethyl-4-(phenylcarbonyl)phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
1DQP
 
 | | CRYSTAL STRUCTURE OF GIARDIA GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH IMMUCILLING | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-1-(S)-(9-DEAZAGUANIN-9-YL)-D-RIBITOL, GUANINE PHOSPHORIBOSYLTRANSFERASE, ISOPROPYL ALCOHOL | | Authors: | Shi, W, Munagala, N.R, Wang, C.C, Li, C.M, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,).
Biochemistry, 39, 2000
|
|
1DT2
 
 | |
1DUA
 
 | |
7X1Z
 
 | | Structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
6MD2
 
 | |
2IS3
 
 | |
7X1Y
 
 | | Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
1XP3
 
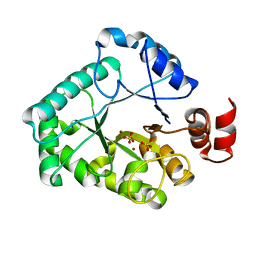 | | Crystal Structure of Endonuclease IV (BA4508) from Bacillus anthracis at 2.57A Resolution. | | Descriptor: | SULFATE ION, ZINC ION, endonuclease IV | | Authors: | Fogg, M.J, Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of Endonuclease IV (BA4508) from Bacillus anthracis at 2.57A Resolution.
To be Published
|
|
3OT7
 
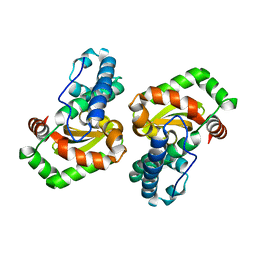 | | Escherichia coli apo-manganese superoxide dismutase | | Descriptor: | Superoxide dismutase [Mn] | | Authors: | Whittaker, M.M, Lerch, T.F, Kirillova, O, Chapman, M.S, Whittaker, J.W. | | Deposit date: | 2010-09-10 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Subunit dissociation and metal binding by Escherichia coli apo-manganese superoxide dismutase.
Arch.Biochem.Biophys., 505, 2011
|
|
7DQB
 
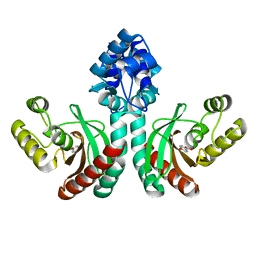 | | Crystal structure of an IclR homolog complexed with 4-hydroxybenzoate from Microbacterium hydrocarbonoxydans in P212121 form | | Descriptor: | IclR homolog, P-HYDROXYBENZOIC ACID | | Authors: | Akiyama, T, Sasaki, Y, Ito, S, Yajima, S. | | Deposit date: | 2020-12-23 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural basis of the conformational changes in Microbacterium hydrocarbonoxydans IclR transcription factor homolog due to ligand binding.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6LX7
 
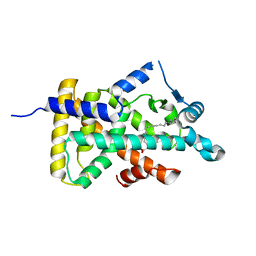 | | X-ray structure of human PPARalpha ligand binding domain-stearic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, STEARIC ACID | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXC
 
 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by delipidation and cross-seeding | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6F2L
 
 | |
6MFR
 
 | |
1DVE
 
 | | CRYSTAL STRUCTURE OF RAT HEME OXYGENASE-1 IN COMPLEX WITH HEME | | Descriptor: | HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Omata, Y, Kakuta, Y, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2000-01-20 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of rat heme oxygenase-1 in complex with heme.
FEBS Lett., 471, 2000
|
|
5UZY
 
 | |
1DQN
 
 | | CRYSTAL STRUCTURE OF GIARDIA GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH A TRANSITION STATE ANALOGUE | | Descriptor: | GUANINE PHOSPHORIBOSYLTRANSFERASE, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Shi, W, Munagala, N.R, Wang, C.C, Li, C.M, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,).
Biochemistry, 39, 2000
|
|
6FO9
 
 | | Vitamin D nuclear receptor complex 2 | | Descriptor: | (1~{R},3~{S},5~{Z})-4-methylidene-5-[(~{E})-3-[3-(6-methyl-6-oxidanyl-heptyl)phenyl]hex-2-enylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2018-02-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Aromatic-Based Design of Highly Active and Noncalcemic Vitamin D Receptor Agonists.
J. Med. Chem., 61, 2018
|
|
6FOD
 
 | | Vitamin D nuclear receptor complex 1 | | Descriptor: | (1~{R},3~{S},5~{Z})-4-methylidene-5-[(~{E})-3-[3-(6-methyl-6-oxidanyl-heptyl)phenyl]pent-2-enylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aromatic-Based Design of Highly Active and Noncalcemic Vitamin D Receptor Agonists.
J. Med. Chem., 61, 2018
|
|
6MFN
 
 | |
