4A0B
 
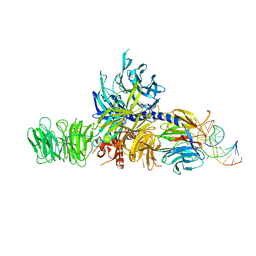 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.8 A resolution (CPD 4) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*DGP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', DNA DAMAGE-BINDING PROTEIN 1, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4BYI
 
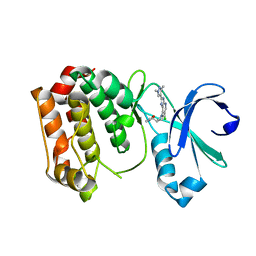 | | Aurora A kinase bound to a highly selective imidazopyridine inhibitor | | Descriptor: | (S)-N-((1-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)pyrrolidin-3-yl)methyl)acetamide, AURORA KINASE A | | Authors: | Joshi, A, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aurora Isoform Selectivity: Design and Synthesis of Imidazo[4,5-B]Pyridine Derivatives as Highly Selective Inhibitors of Aurora-A Kinase in Cells.
J.Med.Chem., 56, 2013
|
|
5M39
 
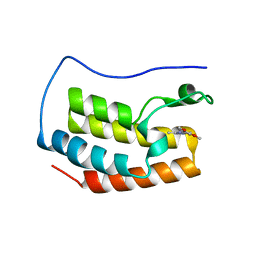 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 1 | | Descriptor: | 6-(3,4-dimethoxyphenyl)-3-methyl-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
5LQ2
 
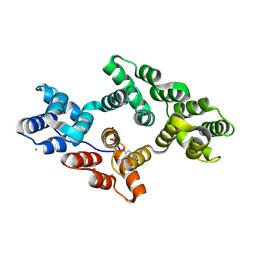 | | Crystal structure of Tyr24 phosphorylated Annexin A2 at 3.4 A resolution | | Descriptor: | Annexin A2, CALCIUM ION | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
4A0L
 
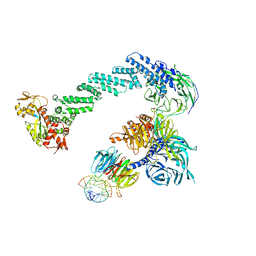 | | Structure of DDB1-DDB2-CUL4B-RBX1 bound to a 12 bp abasic site containing DNA-duplex | | Descriptor: | 12 BP DNA DUPLEX, 12 BP THF CONTAINING DNA DUPLEX, CULLIN-4B, ... | | Authors: | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (7.4 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4CET
 
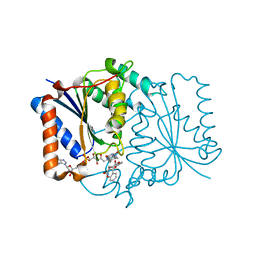 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with dicoumarol at 2.2 A resolution | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
4CF6
 
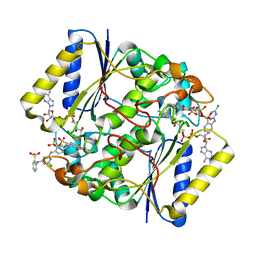 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with Cibacron blue at 2.7 A resolution | | Descriptor: | CIBACRON BLUE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
4A90
 
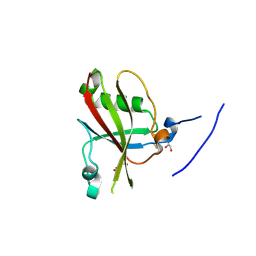 | | Crystal structure of mouse SAP18 residues 1-143 | | Descriptor: | GLYCEROL, HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18 | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A08
 
 | | Structure of hsDDB1-drDDB2 bound to a 13 bp CPD-duplex (purine at D-1 position) at 3.0 A resolution (CPD 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*CP*GP*CP*GP*AP*(TTD)P*GP*CP*GP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*CP*GP*CP*CP*CP*TP*CP*GP*CP*G)-3', ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4CCM
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-1) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4AV1
 
 | | Crystal structure of the human PARP-1 DNA binding domain in complex with DNA | | Descriptor: | 5'-D(*AP*AP*GP*TP*GP*TP*TP*GP*CP*AP*TP*TP)-3', 5'-D(*TP*AP*AP*TP*GP*CP*AP*AP*CP*AP*CP*TP)-3', POLY [ADP-RIBOSE] POLYMERASE 1, ... | | Authors: | Ali, A.A.E, Timinszky, G, Arribas-Bosacoma, R, Kozlowski, M, Hassa, P.O, Hassler, M, Ladurner, A.G, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Zinc-Finger Domains of Parp1 Cooperate to Recognise DNA Strand-Breaks
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EZ8
 
 | | Crystal structure of mouse thymidylate sythase in ternary complex with N(4)-hydroxy-2'-deoxycytidine-5'-monophosphate and the cofactor product, dihydrofolate | | Descriptor: | 2'-deoxy-N-hydroxycytidine 5'-(dihydrogen phosphate), DIHYDROFOLIC ACID, GLYCEROL, ... | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W, Wilk, P, Kierdaszuk, B, Banaszak, K, Gorecka, K, Rode, W. | | Deposit date: | 2012-05-02 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal structures of complexes of mouse thymidylate synthase
crystallized with N4-OH-dCMP alone or in the presence of
N5,10-methylenetetrahydrofolate
Pteridines, 2013
|
|
5NNE
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated TOP2A peptide (K1201ac/K1204ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GKA(ALY)GK(ALY)TQMY | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5NOT
 
 | | Structure of cyclophilin A in complex with 4-chloropyrimidin-5-amine | | Descriptor: | 4-chloranylpyrimidin-5-amine, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOX
 
 | | Structure of cyclophilin A in complex with 2-chloropyridin-3-amine | | Descriptor: | 2-chloranylpyridin-3-amine, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NQQ
 
 | |
3ZZZ
 
 | |
4CZZ
 
 | | Histone demethylase LSD1(KDM1A)-CoREST3 Complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 3 | | Authors: | Barrios, A.P, Gomez, A.V, Saez, J.E, Ciossani, G, Toffolo, E, Battaglioli, E, Mattevi, A, Andres, M.E. | | Deposit date: | 2014-04-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential Properties of Transcriptional Complexes Formed by the Corest Family.
Mol.Cell.Biol., 34, 2014
|
|
4MD9
 
 | |
4MHG
 
 | | Crystal structure of ETV6 bound to a specific DNA sequence | | Descriptor: | Complementary Specific 14 bp DNA, Specific 14 bp DNA, Transcription factor ETV6 | | Authors: | Chan, A.C, De, S, Coyne III, H.J, Okon, M, Murphy, M.E, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2013-08-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
4D2G
 
 | | Crystal structure of human PCNA in complex with p15 peptide | | Descriptor: | P15, PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | DeBiasio, A, Ibanez, A, Mortuza, G, Molina, R, Cordeiro, T.N, Castillo, F, Villate, M, Merino, N, Lelli, M, Diercks, T, Luque, I, Bernardo, P, Montoya, G, Blanco, F.J. | | Deposit date: | 2014-05-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of P15(Paf)-PCNA Complex and Implications for Clamp Sliding During DNA Replication and Repair.
Nat.Commun., 6, 2015
|
|
4AT9
 
 | |
4CI1
 
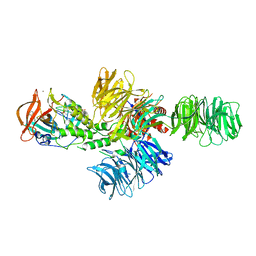 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to thalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Thalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4O78
 
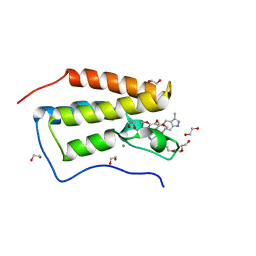 | | Crystal structure of the first bromodomain of human BRD4 in complex with GW612286X | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O9I
 
 | |
