6LXE
 
 | | DROSHA-DGCR8 complex | | Descriptor: | Microprocessor complex subunit DGCR8, Ribonuclease 3, ZINC ION | | Authors: | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
7B0Y
 
 | | Structure of a transcribing RNA polymerase II-U1 snRNP complex | | Descriptor: | 145-nt RNA, DNA-directed RNA polymerase II subunit D, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Zhang, S, Aibara, S, Vos, S.M, Agafonov, D.E, Luehrmann, R, Cramer, P. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science, 371, 2021
|
|
4L8R
 
 | | Structure of mrna stem-loop, human stem-loop binding protein and 3'hexo ternary complex | | Descriptor: | 3'-5' exoribonuclease 1, HISTONE MRNA STEM-LOOP, Histone RNA hairpin-binding protein | | Authors: | Tan, D, Tong, L. | | Deposit date: | 2013-06-17 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Histone Mrna Stem-Loop, Human Stem-Loop Binding Protein, and 3'Hexo Ternary Complex.
Science, 339, 2013
|
|
7DNT
 
 | | mRNA-decapping enzyme g5Rp | | Descriptor: | mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
9FIA
 
 | | SSU(body) structure derived from the SSU sample of the mitoribosome from T. gondii. | | Descriptor: | 30S ribosomal protein S12, putative, 30S ribosomal protein S5, ... | | Authors: | Rocha, R.E.O, Barua, S, Boissier, F, Nguyen, T.T, Hashem, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Apicomplexan mitoribosome from highly fragmented rRNAs to a functional machine.
Nat Commun, 15, 2024
|
|
7U0R
 
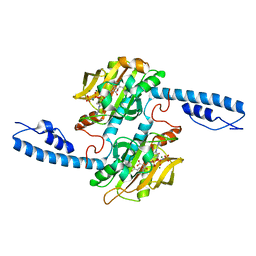 | | Crystal structure of Methanomethylophilus alvus PylRS(N166A/V168A) complexed with meta-trifluoromethyl-2-benzylmalonate and AMP-PNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Swenson, C.V, Roe, L.T, Fricke, R.C.B, Smaga, S.S, Gee, C.L, Schepartz, A. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expanding the substrate scope of pyrrolysyl-transfer RNA synthetase enzymes to include non-alpha-amino acids in vitro and in vivo.
Nat.Chem., 15, 2023
|
|
8UP5
 
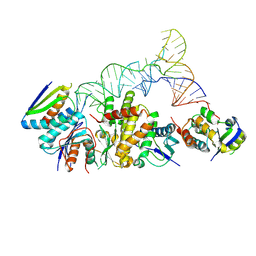 | | Structure of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1) bound to tRNA in its native-like conformation. | | Descriptor: | KEOPS complex subunit Pcc1, Probable bifunctional tRNA threonylcarbamoyladenosine biosynthesis protein, Regulatory protein Cgi121, ... | | Authors: | Ona Chuquimarca, S.M, Beenstock, J, Daou, S, Keszei, A.F.A, Yin, Z, Sicheri, F. | | Deposit date: | 2023-10-21 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structures of KEOPS bound to tRNA reveal functional roles of the kinase Bud32.
Nat Commun, 15, 2024
|
|
5Z98
 
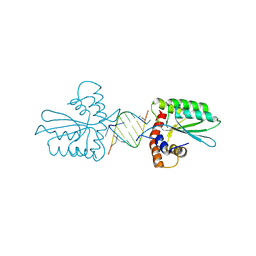 | | Crystal Structure of the Primate APOBEC3H Dimer mediated by RNA Duplex | | Descriptor: | Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like protein 3H, RNA (5'-R(*AP*UP*AP*CP*CP*CP*GP*GP*CP*A)-3'), RNA (5'-R(P*CP*UP*GP*CP*CP*GP*GP*GP*UP*A)-3'), ... | | Authors: | Matsuoka, T, Nagae, T, Ode, H, Watanabe, N, Iwatani, Y. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA.
Nucleic Acids Res., 46, 2018
|
|
1A3M
 
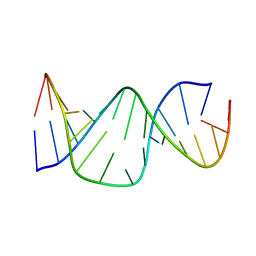 | | PAROMOMYCIN BINDING INDUCES A LOCAL CONFORMATIONAL CHANGE IN THE A SITE OF 16S RRNA, NMR, 20 STRUCTURES | | Descriptor: | 16S RRNA (5'-R(*GP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*UP*UP*C)-3'), 16S RRNA (5'-R(*GP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*C)-3') | | Authors: | Fourmy, D, Yoshizawa, S, Puglisi, J.D. | | Deposit date: | 1998-01-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA.
J.Mol.Biol., 277, 1998
|
|
9D85
 
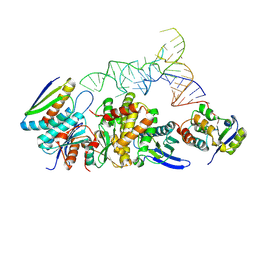 | | Structure of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1) bound to tRNA in a distorted tRNA conformation | | Descriptor: | KEOPS complex subunit Pcc1, Probable bifunctional tRNA threonylcarbamoyladenosine biosynthesis protein, Regulatory protein Cgi121, ... | | Authors: | Ona Chuquimarca, S.M, Beenstock, J, Daou, S, Keszei, A.F.A, Yin, Z, Sicheri, F. | | Deposit date: | 2024-08-18 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structures of KEOPS bound to tRNA reveal functional roles of the kinase Bud32.
Nat Commun, 15, 2024
|
|
1XMQ
 
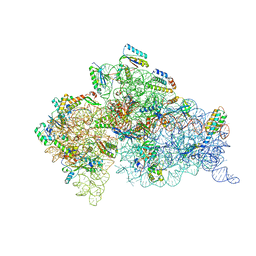 | | Crystal Structure of t6A37-ASLLysUUU AAA-mRNA Bound to the Decoding Center | | Descriptor: | 16s ribosomal RNA, 30S Ribosomal Protein S10, 30S Ribosomal Protein S11, ... | | Authors: | Murphy, F.V, Ramakrishnan, V, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of modifications in codon discrimination by tRNA(Lys)(UUU).
Nat.Struct.Mol.Biol., 11, 2004
|
|
4U38
 
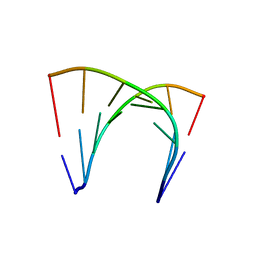 | | RNA duplex containing UU mispair | | Descriptor: | RNA (5'-R(*GP*GP*UP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
7C45
 
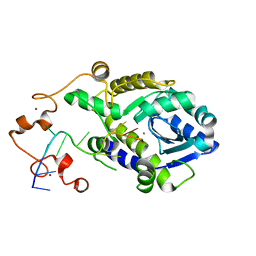 | |
4OOG
 
 | |
3TRA
 
 | |
3NNH
 
 | | Crystal Structure of the CUGBP1 RRM1 with GUUGUUUUGUUU RNA | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7501 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
1Y1W
 
 | | Complete RNA Polymerase II elongation complex | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*T)-3', 5'-D(P*AP*GP*TP*AP*CP*TP*TP*AP*CP*GP*CP*CP*TP*GP*GP*TP*CP*AP*T)-3', 5'-R(*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Cramer, P, Kettenberger, H, Armache, K.-J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Complete RNA Polymerase II Elongation Complex Structure and Its Interactions with NTP and TFIIS
Mol.Cell, 16, 2004
|
|
1Y77
 
 | | Complete RNA Polymerase II elongation complex with substrate analogue GMPCPP | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*T)-3', 5'-D(P*AP*GP*TP*AP*CP*TP*TP*AP*CP*T*CP*GP*CP*CP*TP*GP*GP*TP*CP*TP*G)-3', 5'-R(*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Kettenberger, H, Armache, K.-J, Cramer, P. | | Deposit date: | 2004-12-08 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Complete RNA Polymerase II Elongation Complex Structure and Its Interactions with NTP and TFIIS
Mol.Cell, 16, 2004
|
|
3S7T
 
 | |
4BY9
 
 | | The structure of the Box CD enzyme reveals regulation of rRNA methylation | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*CP*AP*UP*CP*AP*CP)-3', 50S RIBOSOMAL PROTEIN L7AE, FIBRILLARIN-LIKE RRNA/TRNA 2'-O-METHYLTRANSFERASE, ... | | Authors: | Lapinaite, A, Simon, B, Skjaerven, L, Rakwalska-Bange, M, Gabel, F, Carlomagno, T. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Box C/D Enzyme Reveals Regulation of RNA Methylation.
Nature, 502, 2013
|
|
7VKI
 
 | | ESRP1 qRRM2 in complex with 12mer-RNA | | Descriptor: | Epithelial splicing regulatory protein 1, RNA (12-mer) | | Authors: | Wu, B.X, Patel, D.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ESRP1 controls biogenesis and function of a large abundant multiexon circRNA.
Nucleic Acids Res., 2023
|
|
6M8Q
 
 | | Cleavage and Polyadenylation Specificity Factor Subunit 3 (CPSF3) in complex with NVP-LTM531 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 3, N-{3,5-dichloro-2-hydroxy-4-[2-(4-methylpiperazin-1-yl)ethoxy]benzene-1-carbonyl}-L-phenylalanine, PHOSPHATE ION, ... | | Authors: | Weihofen, W.A, Salcius, M, Michaud, G. | | Deposit date: | 2018-08-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing's sarcoma.
Nat.Chem.Biol., 16, 2020
|
|
4V9E
 
 | |
8Q3B
 
 | | The closed state of the ASFV apo-RNA polymerase | | Descriptor: | DNA-directed RNA polymerase RPB1 homolog, DNA-directed RNA polymerase RPB10 homolog, DNA-directed RNA polymerase RPB2 homolog, ... | | Authors: | Pilotto, S, Sykora, M, Cackett, G, Werner, F. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the recombinant RNA polymerase from African Swine Fever Virus.
Nat Commun, 15, 2024
|
|
8Q3K
 
 | | The open state of the ASFV apo-RNA polymerase | | Descriptor: | DNA-directed RNA polymerase RPB1 homolog, DNA-directed RNA polymerase RPB10 homolog, DNA-directed RNA polymerase RPB2 homolog, ... | | Authors: | Pilotto, S, Sykora, M, Cackett, G, Werner, F. | | Deposit date: | 2023-08-04 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structure of the recombinant RNA polymerase from African Swine Fever Virus.
Nat Commun, 15, 2024
|
|
